

You can:
| Name | Cannabinoid receptor 1 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | CNR1 |
| Synonym | CB1 Central cannabinoid receptor SKR6R THC receptor CB1R [ Show all ] |
| Disease | Obesity; Diabetes Chemotherapy-induced nausea Diabetes; Obesity Drug abuse Hypertension; Diabetes; Obesity [ Show all ] |
| Length | 472 |
| Amino acid sequence | MKSILDGLADTTFRTITTDLLYVGSNDIQYEDIKGDMASKLGYFPQKFPLTSFRGSPFQEKMTAGDNPQLVPADQVNITEFYNKSLSSFKENEENIQCGENFMDIECFMVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCRPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRKDSRNVFLFKLGGVTASFTASVGSLFLTAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCEKLQSVCSDIFPHIDETYLMFWIGVTSVLLLFIVYAYMYILWKAHSHAVRMIQRGTQKSIIIHTSEDGKVQVTRPDQARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMFPSCEGTAQPLDNSMGDSDCLHKHANNAASVHRAAESCIKSTVKIAKVTMSVSTDTSAEAL |
| UniProt | P21554 |
| Protein Data Bank | 5tjv, 5u09, 5xr8, 5xra, 6n4b, 5tgz |
| GPCR-HGmod model | P21554 |
| 3D structure model | This structure is from PDB ID 5tjv. |
| BioLiP | BL0384680, BL0364157, BL0384679, BL0384681, BL0384682, BL0384683, BL0384684, BL0440253, BL0440254,BL0440255, BL0363267, BL0361447, BL0361446 |
| Therapeutic Target Database | T76685 |
| ChEMBL | CHEMBL218 |
| IUPHAR | 56 |
| DrugBank | BE0000061 |
| Name | SCHEMBL604437 |
|---|---|
| Molecular formula | C18H25N5O |
| IUPAC name | (7aS,11aS)-4-piperazin-1-yl-5,6,7a,8,9,10,11,11a-octahydro-[1]benzofuro[2,3-h]quinazolin-2-amine |
| Molecular weight | 327.432 |
| Hydrogen bond acceptor | 6 |
| Hydrogen bond donor | 2 |
| XlogP | 1.3 |
| Synonyms | AKOS024457726 UNII-6BVK16R925 component DJKJVWJQAVGLHJ-YPMHNXCESA-N (+/-)-(7aR*,11aR*)-5,6,7a,8,9,10,11,11a-Octahydro-4-(1-piperazinyl)-benzofuran[2,3-h]quinazolin-2-amine CHEMBL519240 (12S,17S)-6-(piperazin-1-yl)-11-oxa-3,5-diazatetracyclo[8.7.0.0^{2,7}.0^{12,17}]heptadeca-1(10),2(7),3,5-tetraen-4-amine [ Show all ] |
| Inchi Key | DJKJVWJQAVGLHJ-YPMHNXCESA-N |
| Inchi ID | InChI=1S/C18H25N5O/c19-18-21-16-12(17(22-18)23-9-7-20-8-10-23)5-6-14-15(16)11-3-1-2-4-13(11)24-14/h11,13,20H,1-10H2,(H2,19,21,22)/t11-,13+/m1/s1 |
| PubChem CID | 25109291 |
| ChEMBL | CHEMBL519240 |
| IUPHAR | N/A |
| BindingDB | 26226 |
| DrugBank | N/A |
Structure | 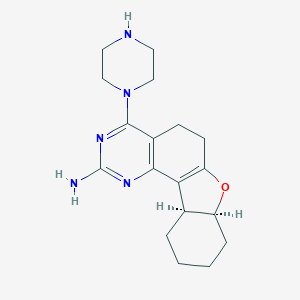 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| Activity | 82.6 % | PMID18983139 | ChEMBL |
| Inhibition | 17.0 % | PMID18983139 | ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417