

You can:
| Name | P2Y purinoceptor 2 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | P2RY2 |
| Synonym | Purinergic receptor purinergic receptor P2Y P2Y2 receptor P2Y2 P2Y purinoceptor 2 [ Show all ] |
| Disease | Dry eye disease Constipation Cystic fibrosis Lung cancer |
| Length | 377 |
| Amino acid sequence | MAADLGPWNDTINGTWDGDELGYRCRFNEDFKYVLLPVSYGVVCVPGLCLNAVALYIFLCRLKTWNASTTYMFHLAVSDALYAASLPLLVYYYARGDHWPFSTVLCKLVRFLFYTNLYCSILFLTCISVHRCLGVLRPLRSLRWGRARYARRVAGAVWVLVLACQAPVLYFVTTSARGGRVTCHDTSAPELFSRFVAYSSVMLGLLFAVPFAVILVCYVLMARRLLKPAYGTSGGLPRAKRKSVRTIAVVLAVFALCFLPFHVTRTLYYSFRSLDLSCHTLNAINMAYKVTRPLASANSCLDPVLYFLAGQRLVRFARDAKPPTGPSPATPARRRLGLRRSDRTDMQRIEDVLGSSEDSRRTESTPAGSENTKDIRL |
| UniProt | P41231 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | P41231 |
| 3D structure model | This predicted structure model is from GPCR-EXP P41231. |
| BioLiP | N/A |
| Therapeutic Target Database | T93515 |
| ChEMBL | CHEMBL4398 |
| IUPHAR | 324 |
| DrugBank | BE0002401 |
| Name | Adenosine triphosphate |
|---|---|
| Molecular formula | C10H16N5O13P3 |
| IUPAC name | [[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy-hydroxyphosphoryl] phosphono hydrogen phosphate |
| Molecular weight | 507.181 |
| Hydrogen bond acceptor | 17 |
| Hydrogen bond donor | 7 |
| XlogP | -5.7 |
| Synonyms | H4atp Adenosine 5 inverted exclamation marka-triphosphate (ATP) disodium salt hydrate SCHEMBL8979 Adenosine 5'-triphosphate (intravenous, paroxysmal supraventricular tachycardia (PSVT)), Duska Triphosphoric acid adenosine ester [ Show all ] |
| Inchi Key | ZKHQWZAMYRWXGA-KQYNXXCUSA-N |
| Inchi ID | InChI=1S/C10H16N5O13P3/c11-8-5-9(13-2-12-8)15(3-14-5)10-7(17)6(16)4(26-10)1-25-30(21,22)28-31(23,24)27-29(18,19)20/h2-4,6-7,10,16-17H,1H2,(H,21,22)(H,23,24)(H2,11,12,13)(H2,18,19,20)/t4-,6-,7-,10-/m1/s1 |
| PubChem CID | 5957 |
| ChEMBL | CHEMBL14249 |
| IUPHAR | 1713 |
| BindingDB | 2, 50366480 |
| DrugBank | DB00171 |
Structure | 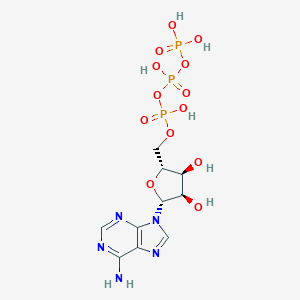 |
| Lipinski's druglikeness | This ligand has more than 5 hydrogen bond donor. This ligand has more than 10 hydrogen bond acceptor. This ligand is heavier than 500 daltons. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| EC50 | <300000.0 nM | PMID19419204 | ChEMBL |
| EC50 | 30.1 nM | PMID19419204 | ChEMBL |
| EC50 | 63.1 nM | PMID19419204 | BindingDB,ChEMBL |
| EC50 | 79.4328 - 229.087 nM | PMID8564228, PMID11754592, PMID12213051 | IUPHAR |
| EC50 | 85.0 nM | PMID11985476, PMID19419868, PMID17302398 | BindingDB,ChEMBL |
| EC50 | 95.8 nM | PMID19419204 | BindingDB,ChEMBL |
| EC50 | 111.0 nM | PMID19419204 | ChEMBL |
| EC50 | 120.0 nM | PMID19419204 | ChEMBL |
| EC50 | 183.0 nM | PMID19419204 | ChEMBL |
| EC50 | 230.0 nM | PMID12213051 | BindingDB,ChEMBL |
| EC50 | 237.0 nM | PMID19419204 | ChEMBL |
| EC50 | 239.0 nM | PMID19419204 | ChEMBL |
| EC50 | 299.0 nM | PMID19419204 | ChEMBL |
| EC50 | 1000.0 nM | PMID19419204 | ChEMBL |
| EC50 | 11700.0 nM | PMID19419204 | ChEMBL |
| Efficacy | 5.0 % | PMID19419204 | ChEMBL |
| Efficacy | 41.0 % | PMID19419204 | ChEMBL |
| Efficacy | 87.0 % | PMID19419204 | ChEMBL |
| Efficacy | 92.0 % | PMID19419204 | ChEMBL |
| Efficacy | 97.0 % | PMID19419204 | ChEMBL |
| Efficacy | 104.0 % | PMID19419204 | ChEMBL |
| Efficacy | 110.0 % | PMID19419204 | ChEMBL |
| Efficacy | 115.0 % | PMID19419204 | ChEMBL |
| Efficacy | 117.0 % | PMID19419204 | ChEMBL |
| Efficacy | 126.0 % | PMID19419204 | ChEMBL |
| Efficacy | 130.0 % | PMID19419204 | ChEMBL |
| Ratio EC50 | 2.0 - | PMID19419204 | ChEMBL |
| Ratio EC50 | 3.0 - | PMID19419204 | ChEMBL |
| Ratio EC50 | 4.0 - | PMID19419204 | ChEMBL |
| Ratio EC50 | 10.0 - | PMID19419204 | ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417