

You can:
| Name | 5-hydroxytryptamine receptor 1B |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Htr1b |
| Synonym | HTR1D2 5-hydroxytryptamine (serotonin) receptor 1B, G protein-coupled 5-HT1Dbeta 5-HT1DB 5-HT1B serotonin receptor [ Show all ] |
| Disease | N/A for non-human GPCRs |
| Length | 386 |
| Amino acid sequence | MEEQGIQCAPPPPATSQTGVPLANLSHNCSADDYIYQDSIALPWKVLLVALLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIMHLCVIALDRYWAITDAVDYSAKRTPKRAAIMIVLVWVFSISISLPPFFWRQAKAEEEVLDCFVNTDHVLYTVYSTVGAFYLPTLLLIALYGRIYVEARSRILKQTPNKTGKRLTRAQLITDSPGSTSSVTSINSRVPEVPSESGSPVYVNQVKVRVSDALLEKKKLMAARERKATKTLGIILGAFIVCWLPFFIISLVMPICKDACWFHMAIFDFFNWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFKCTG |
| UniProt | P28564 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL3459 |
| IUPHAR | 2 |
| DrugBank | N/A |
| Name | quipazine |
|---|---|
| Molecular formula | C13H15N3 |
| IUPAC name | 2-piperazin-1-ylquinoline |
| Molecular weight | 213.284 |
| Hydrogen bond acceptor | 3 |
| Hydrogen bond donor | 1 |
| XlogP | 1.5 |
| Synonyms | KBio2_007349 LS-142110 NCGC00015872-06 2-Piperazin-1-yl-quinoline (Quipazine) Prestwick0_000617 [ Show all ] |
| Inchi Key | XRXDAJYKGWNHTQ-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C13H15N3/c1-2-4-12-11(3-1)5-6-13(15-12)16-9-7-14-8-10-16/h1-6,14H,7-10H2 |
| PubChem CID | 5011 |
| ChEMBL | CHEMBL18772 |
| IUPHAR | 173 |
| BindingDB | 50014407 |
| DrugBank | N/A |
Structure | 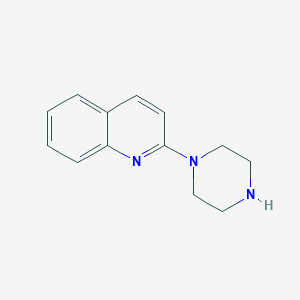 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| IC50 | 380.0 nM | PMID9357534 | BindingDB,ChEMBL |
| IC50 | 7079.46 nM | PMID8642566, PMID9191957 | BindingDB,ChEMBL |
| Ki | 199.53 nM | PMID10543880 | ChEMBL |
| Ki | 200.0 nM | PMID2583244 | BindingDB |
| Ki | 208.0 nM | PMID7629808 | BindingDB,ChEMBL |
| Ki | 249.0 nM | PMID1738111 | BindingDB |
| Ki | 310.0 nM | PMID9513601, PMID10229626 | BindingDB,ChEMBL |
| Ki | 320.0 nM | PMID3543362 | BindingDB,ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417