

You can:
| Name | Kappa-type opioid receptor |
|---|---|
| Species | Cavia porcellus (Guinea pig) |
| Gene | OPRK1 |
| Synonym | K-OR-1 KOR-1 |
| Disease | N/A for non-human GPCRs |
| Length | 380 |
| Amino acid sequence | MGRRRQGPAQPASELPARNACLLPNGSAWLPGWAEPDGNGSAGPQDEQLEPAHISPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVISIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIILGGTKVREDVDIIECSLQFPDDDYSWWDLFMKICVFVFAFVIPVLIIIVCYTLMILRLKSVRLLSGSREKDRNLRRITRLVLVVVAVFIICWTPIHIFILVEALGSTSHSTAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFPIKMRMERQSTSRVRNTVQDPAYMRNVDGVNKPV |
| UniProt | P41144 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL3952 |
| IUPHAR | N/A |
| DrugBank | N/A |
| Name | U50488 |
|---|---|
| Molecular formula | C19H26Cl2N2O |
| IUPAC name | 2-(3,4-dichlorophenyl)-N-methyl-N-[(1R,2R)-2-pyrrolidin-1-ylcyclohexyl]acetamide |
| Molecular weight | 369.33 |
| Hydrogen bond acceptor | 2 |
| Hydrogen bond donor | 0 |
| XlogP | 4.4 |
| Synonyms | Benzeneacetamide, 3,4-dichloro-N-methyl-N-(2-(1-pyrrolidinyl)cyclohexyl)-, trans- Lopac-U-110 SCHEMBL332193 U-50488H 3,4-Dichloro-N-methyl-N-(2-(1-pyrrolidinyl)-cyclohexyl)-benzeneacetamide, (trans)-Isomer [ Show all ] |
| Inchi Key | VQLPLYSROCPWFF-QZTJIDSGSA-N |
| Inchi ID | InChI=1S/C19H26Cl2N2O/c1-22(19(24)13-14-8-9-15(20)16(21)12-14)17-6-2-3-7-18(17)23-10-4-5-11-23/h8-9,12,17-18H,2-7,10-11,13H2,1H3/t17-,18-/m1/s1 |
| PubChem CID | 3036289 |
| ChEMBL | CHEMBL441765 |
| IUPHAR | 1652 |
| BindingDB | 50000296 |
| DrugBank | N/A |
Structure | 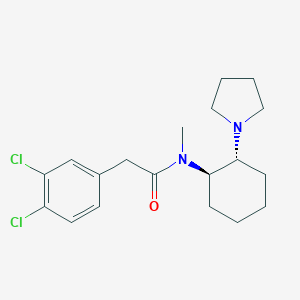 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| Control | 38.0 % | PMID1319495 | ChEMBL |
| Control | 105.0 % | PMID1319495 | ChEMBL |
| Control | 109.0 % | PMID1319495 | ChEMBL |
| IC50 | 0.303 nM | PMID18990576 | BindingDB,ChEMBL |
| IC50 | 1.12 nM | PMID18829333 | BindingDB,ChEMBL |
| IC50 | 1.14 nM | PMID18337104 | BindingDB,ChEMBL |
| IC50 | 95.5 nM | PMID1659636 | ChEMBL |
| IC50 | 96.0 nM | PMID1846918, PMID1659636 | BindingDB,ChEMBL |
| Ke | 0.031 nM | PMID18829333 | ChEMBL |
| Ki | 0.31 nM | PMID20441176 | BindingDB |
| Ki | 0.31 nM | PMID20441176, PMID21481987 | ChEMBL |
| Ki | 0.34 nM | PMID24856182, PMID25062506 | BindingDB |
| Ki | 0.34 nM | PMID24856182, PMID25062506 | ChEMBL |
| Ki | 0.36 nM | PMID11591513, PMID10633042 | BindingDB |
| Ki | 0.36 nM | PMID11591513, PMID10633042 | ChEMBL |
| Ki | 0.49 nM | PMID17420073, PMID11495592 | BindingDB,ChEMBL |
| Ki | 0.49 nM | PMID11495592 | BindingDB |
| Ki | 0.89 nM | PMID10841791 | BindingDB,ChEMBL |
| Ki | 0.912 nM | PMID8632410 | ChEMBL |
| Ki | 0.97 nM | PMID1846921 | BindingDB |
| Ki | 0.97 nM | PMID1846921 | ChEMBL |
| Ki | 1.1 nM | PMID20599386 | BindingDB |
| Ki | 1.14 nM | PMID20599386 | ChEMBL |
| Ki | 1.16 nM | PMID1361580 | ChEMBL |
| Ki | 1.2 nM | PMID1361580 | BindingDB |
| Ki | 1.5 nM | PMID25062506 | BindingDB,ChEMBL |
| Ki | 1.67 nM | PMID25147605 | ChEMBL |
| Ki | 1.7 nM | PMID25147605 | BindingDB |
| Ki | 2.2 nM | PMID1652025, PMID1323679 | BindingDB,ChEMBL |
| Ki | 5.0 nM | PMID10956208 | BindingDB |
| Ki | 5.01 nM | PMID10956208 | ChEMBL |
| Ki | 10.0 nM | PMID2832603, PMID1846919 | BindingDB,ChEMBL |
| Ki | 15.0 nM | PMID1319495 | BindingDB,ChEMBL |
| Ki | 16.5 nM | PMID8390575 | ChEMBL |
| Ki | 17.0 nM | PMID8390575 | BindingDB |
| Ki | 20.5 nM | PMID8390575 | ChEMBL |
| Ki | 21.0 nM | PMID8390575 | BindingDB |
| Ki | 109.0 nM | PMID2547074 | BindingDB,ChEMBL |
| Ki | 204.0 nM | PMID8390575 | BindingDB,ChEMBL |
| Ki | 299.0 nM | PMID2547074 | BindingDB,ChEMBL |
| Ki | 1298.0 nM | PMID2547074 | BindingDB,ChEMBL |
| Ratio | 88.0 - | PMID1846919 | ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417