

You can:
| Name | Beta-2 adrenergic receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | ADRB2 |
| Synonym | beta-2 adrenergic receptor Gpcr7 beta2-adrenoceptor Adrb-2 ADRB2R [ Show all ] |
| Disease | Premature labour Premature ejaculation Obesity Neurogenic bladder dysfunction Hypertension [ Show all ] |
| Length | 413 |
| Amino acid sequence | MGQPGNGSAFLLAPNGSHAPDHDVTQERDEVWVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILMKMWTFGNFWCEFWTSIDVLCVTASIETLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWYRATHQEAINCYANETCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLQKIDKSEGRFHVQNLSQVEQDGRTGHGLRRSSKFCLKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQDNLIRKEVYILLNWIGYVNSGFNPLIYCRSPDFRIAFQELLCLRRSSLKAYGNGYSSNGNTGEQSGYHVEQEKENKLLCEDLPGTEDFVGHQGTVPSDNIDSQGRNCSTNDSLL |
| UniProt | P07550 |
| Protein Data Bank | 3nya, 3ny9, 3ny8, 3d4s, 2rh1, 3pds, 4gbr, 4lde, 6mxt, 6csy, 5x7d, 5jqh, 5d6l, 5d5b, 5d5a, 4ldo, 4ldl, 4qkx |
| GPCR-HGmod model | P07550 |
| 3D structure model | This structure is from PDB ID 3nya. |
| BioLiP | BL0257082, BL0113951, BL0113950, BL0257084, BL0257085, BL0283869, BL0333729, BL0333730, BL0333731,BL0333732,BL0333733, BL0333734, BL0333735, BL0333736,BL0333737,BL0333738, BL0113952,BL0113953,BL0113954, BL0147310, BL0257081, BL0257080, BL0232997, BL0192129, BL0257083, BL0185746,BL0185747, BL0185745, BL0185743,BL0185744, BL0185742, BL0185740,BL0185741, BL0147311,BL0147312, BL0351701,BL0351703, BL0351702,BL0351704, BL0192128, BL0433200, BL0433199, BL0430930, BL0430929, BL0388810, BL0388809, BL0354449, BL0185748, BL0354450, BL0354451,BL0354452,BL0354453, BL0388807,BL0388808 |
| Therapeutic Target Database | T24555, T52522 |
| ChEMBL | CHEMBL210 |
| IUPHAR | 29 |
| DrugBank | BE0000694 |
| Name | Inolin |
|---|---|
| Molecular formula | C19H23NO5 |
| IUPAC name | 1-[(3,4,5-trimethoxyphenyl)methyl]-1,2,3,4-tetrahydroisoquinoline-6,7-diol |
| Molecular weight | 345.395 |
| Hydrogen bond acceptor | 6 |
| Hydrogen bond donor | 3 |
| XlogP | 2.5 |
| Synonyms | AQ-110 PDSP1_000588 Tretoquinol, (+/-)- (+/-)-6,7-Dihydroxy-1-(3',4',5'-trimethoxybenzyl)-1,2,3,4-tetrahydroisoquinoline 1-(3,4,5-Trimethoxybenzyl)-6,7-dihydroxy-1,2,3,4-tetrahydroisoquinoline [ Show all ] |
| Inchi Key | RGVPOXRFEPSFGH-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C19H23NO5/c1-23-17-7-11(8-18(24-2)19(17)25-3)6-14-13-10-16(22)15(21)9-12(13)4-5-20-14/h7-10,14,20-22H,4-6H2,1-3H3 |
| PubChem CID | 5581 |
| ChEMBL | CHEMBL299175 |
| IUPHAR | N/A |
| BindingDB | 50019355 |
| DrugBank | N/A |
Structure | 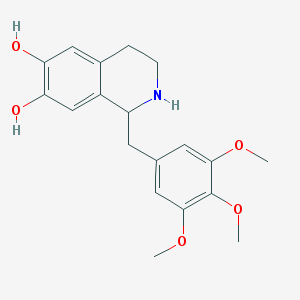 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| Activation | 77.0 % | PMID11055339 | ChEMBL |
| EC50 | 3.5 nM | PMID11055339 | BindingDB,ChEMBL |
| IA | 95.0 - | PMID10691685 | ChEMBL |
| IC50 | 6.1 nM | PMID11055339 | BindingDB,ChEMBL |
| Intrinsic activity | 95.0 - | PMID10377236 | ChEMBL |
| Ki | 20.89 nM | PMID7915318 | BindingDB |
| Ki | 21.37 nM | PMID7915318 | PDSP |
| Ki | 38.01 nM | PMID7915318 | PDSP |
| Ki | 43.65 nM | PMID10377236, PMID8809159 | ChEMBL |
| Ki | 45.7 nM | PMID7915318 | BindingDB |
| pK | 8.33 - | PMID10691685 | ChEMBL |
| pKact | 8.33 - | PMID10377236 | ChEMBL |
| PR | 1.0 - | PMID8809159 | ChEMBL |
| Selectivity ratio | 1.0 - | PMID9016331 | ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417