

You can:
| Name | Melatonin receptor type 1A |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | MTNR1A |
| Synonym | MT1 receptor MelR Mel1a receptor Mel-1A-R |
| Disease | Insomnia Anxiety disorder Circadian rhythm sleep disorder Major depressive disorder Sleep disorders |
| Length | 350 |
| Amino acid sequence | MQGNGSALPNASQPVLRGDGARPSWLASALACVLIFTIVVDILGNLLVILSVYRNKKLRNAGNIFVVSLAVADLVVAIYPYPLVLMSIFNNGWNLGYLHCQVSGFLMGLSVIGSIFNITGIAINRYCYICHSLKYDKLYSSKNSLCYVLLIWLLTLAAVLPNLRAGTLQYDPRIYSCTFAQSVSSAYTIAVVVFHFLVPMIIVIFCYLRIWILVLQVRQRVKPDRKPKLKPQDFRNFVTMFVVFVLFAICWAPLNFIGLAVASDPASMVPRIPEWLFVASYYMAYFNSCLNAIIYGLLNQNFRKEYRRIIVSLCTARVFFVDSSNDVADRVKWKPSPLMTNNNVVKVDSV |
| UniProt | P48039 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | P48039 |
| 3D structure model | This predicted structure model is from GPCR-EXP P48039. |
| BioLiP | N/A |
| Therapeutic Target Database | T97613 |
| ChEMBL | CHEMBL1945 |
| IUPHAR | 287 |
| DrugBank | BE0000515 |
| Name | CHEMBL124783 |
|---|---|
| Molecular formula | C16H22N2O2 |
| IUPAC name | N-[3-(2-propyl-1,3-benzoxazol-7-yl)propyl]propanamide |
| Molecular weight | 274.364 |
| Hydrogen bond acceptor | 3 |
| Hydrogen bond donor | 1 |
| XlogP | 3.4 |
| Synonyms | SCHEMBL6780772 BDBM50149159 N-[3-(2-Propyl-benzooxazol-7-yl)-propyl]-propionamide |
| Inchi Key | BMCYPJHTYPAAIJ-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C16H22N2O2/c1-3-7-15-18-13-10-5-8-12(16(13)20-15)9-6-11-17-14(19)4-2/h5,8,10H,3-4,6-7,9,11H2,1-2H3,(H,17,19) |
| PubChem CID | 22718661 |
| ChEMBL | CHEMBL124783 |
| IUPHAR | N/A |
| BindingDB | 50149159 |
| DrugBank | N/A |
Structure | 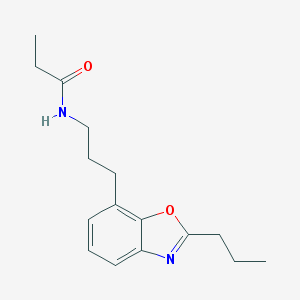 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| Ki | 51.0 nM | PMID15203165 | BindingDB,ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417