

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MQGNGSALPNASQPVLRGDGARPSWLASALACVLIFTIVVDILGNLLVILSVYRNKKLRNAGNIFVVSLAVADLVVAIYPYPLVLMSIFNNGWNLGYLHCQVSGFLMGLSVIGSIFNITGIAINRYCYICHSLKYDKLYSSKNSLCYVLLIWLLTLAAVLPNLRAGTLQYDPRIYSCTFAQSVSSAYTIAVVVFHFLVPMIIVIFCYLRIWILVLQVRQRVKPDRKPKLKPQDFRNFVTMFVVFVLFAICWAPLNFIGLAVASDPASMVPRIPEWLFVASYYMAYFNSCLNAIIYGLLNQNFRKEYRRIIVSLCTARVFFVDSSNDVADRVKWKPSPLMTNNNVVKVDSV | |
| CCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCSSSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCSSSCCCC | |
| 99985899888887777888887499999999999999999999987420146148988498999999999999999998999999998286468737886899999999999999999999981325511575864465887878255999999999999999865784798338985068642799999999999999999999999999998854004753125788988759999999999999998999999999997785123326899999999999999999999999749999999999998864588998888887666765578998678722634779 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MQGNGSALPNASQPVLRGDGARPSWLASALACVLIFTIVVDILGNLLVILSVYRNKKLRNAGNIFVVSLAVADLVVAIYPYPLVLMSIFNNGWNLGYLHCQVSGFLMGLSVIGSIFNITGIAINRYCYICHSLKYDKLYSSKNSLCYVLLIWLLTLAAVLPNLRAGTLQYDPRIYSCTFAQSVSSAYTIAVVVFHFLVPMIIVIFCYLRIWILVLQVRQRVKPDRKPKLKPQDFRNFVTMFVVFVLFAICWAPLNFIGLAVASDPASMVPRIPEWLFVASYYMAYFNSCLNAIIYGLLNQNFRKEYRRIIVSLCTARVFFVDSSNDVADRVKWKPSPLMTNNNVVKVDSV | |
| 36223132323333333442312200000012212301320331320000000003402100000020003012010201110000000263200020011000000000000001000000000010001002134301320000000000210210011110102033454101010213332000000033113310200020011000101334441454544443444200000000000010103111100000000233324330010010200210120002300010001640040013002101344343444644444434345444334444344446 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCSSSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCSSSCCCC MQGNGSALPNASQPVLRGDGARPSWLASALACVLIFTIVVDILGNLLVILSVYRNKKLRNAGNIFVVSLAVADLVVAIYPYPLVLMSIFNNGWNLGYLHCQVSGFLMGLSVIGSIFNITGIAINRYCYICHSLKYDKLYSSKNSLCYVLLIWLLTLAAVLPNLRAGTLQYDPRIYSCTFAQSVSSAYTIAVVVFHFLVPMIIVIFCYLRIWILVLQVRQRVKPDRKPKLKPQDFRNFVTMFVVFVLFAICWAPLNFIGLAVASDPASMVPRIPEWLFVASYYMAYFNSCLNAIIYGLLNQNFRKEYRRIIVSLCTARVFFVDSSNDVADRVKWKPSPLMTNNNVVKVDSV | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.23 | 0.24 | 0.85 | 3.00 | Download | --------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLM--TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSPTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS---KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG----------------------------------- | |||||||||||||||||||
| 2 | 3rzeA | 0.26 | 0.21 | 0.75 | 4.06 | Download | ----------------------------PLVVVLSTICLVTVGLNLLVLYAVRSERKLHTVGNLYIVSLSVADLIVGAVVMPMNILYLLMSKWSLGRPLCLFWLSMDYVASTASIFSVFILCIDRYRSVQQPLRYLKYRTKTRASATILGAWFLSFLWVIPILG----WNHRREDKCETDFYDVTWFKVMTAIINFYLPTLLMLWFYAKIYKAVRHM-------------NRERKAAKQLGFIMAAFILCWIPYFIFFMVIAFCKNCCNEH----LHMFTIWLGYINSTLNPLIYPLCNENFKKTFKRIL---------------------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.23 | 0.24 | 0.85 | 3.83 | Download | --------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATST-LPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLS---GSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG----------------------------------- | |||||||||||||||||||
| 4 | 4ib4 | 0.24 | 0.25 | 0.84 | 1.55 | Download | -----------------EEQGNK---LHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMEAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIKGIET-NPNNITCVLTKERFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSC-NQTTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR----------------------------------- | |||||||||||||||||||
| 5 | 3uon | 0.23 | 0.22 | 0.81 | 1.18 | Download | -----------------------TFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIVTVEDGECYIQFFSNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIFGGSAAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPC----IPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM--------------------------------------- | |||||||||||||||||||
| 6 | 4n6hA | 0.23 | 0.24 | 0.84 | 3.24 | Download | -----------------ARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALA-TSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS---KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG----------------------------------- | |||||||||||||||||||
| 7 | 3uon | 0.23 | 0.22 | 0.80 | 1.70 | Download | --------------------------VVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIVTVEDGECYIQFFSNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIFGAADAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPC----IPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM--------------------------------------- | |||||||||||||||||||
| 8 | 4n6hA | 0.23 | 0.24 | 0.84 | 4.42 | Download | ----------------GSPGASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATST-LPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSPTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS---KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG----------------------------------- | |||||||||||||||||||
| 9 | 4iaqA | 0.24 | 0.21 | 0.79 | 3.18 | Download | ---------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFFW------RQASECVVNTD-HILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRII----QKYLLMAARERKATKTLGIILGAFIVCWLPFFIISLVMPIH---------LAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK------------------------------------- | |||||||||||||||||||
| 10 | 5c1mA | 0.24 | 0.23 | 0.83 | 3.94 | Download | -----------GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVRYTKMKTATNIYIFNLALADALATST-LPFQSVNYLMGTWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRTPRNAKIVNVCNWILSSAIGLPVMFMATTKYRQGSIDCTLTFSHPNLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMLSGS---KEKDRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKALITIPETT-FQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF-------------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
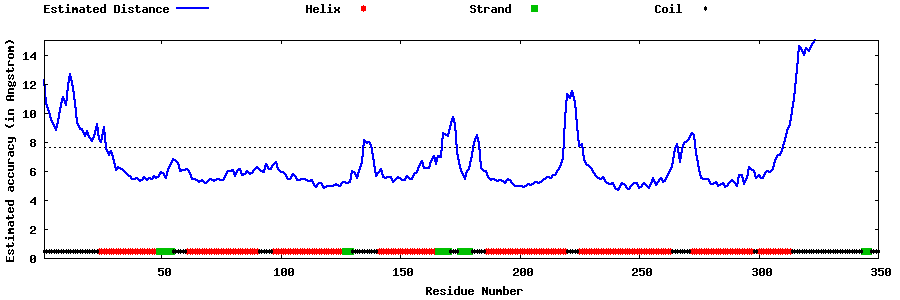
| Generated 3D models | Estimated local accuracy of models | ||
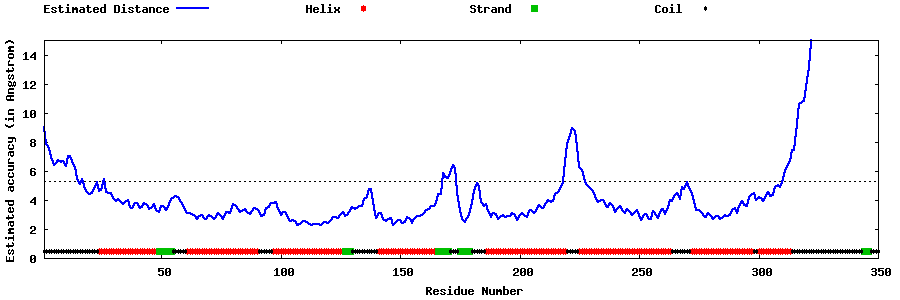 | |||
|
|
|||
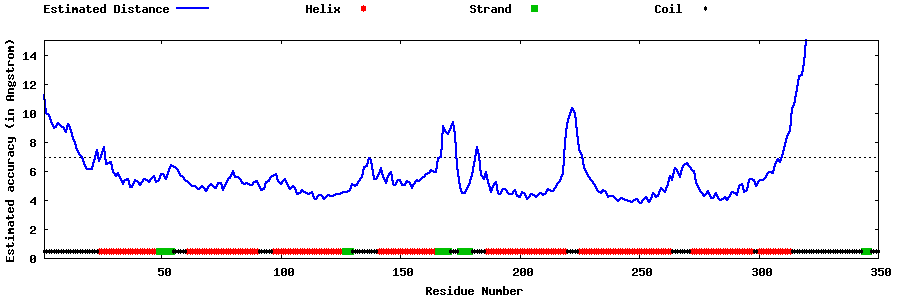 | |||
|
| |||
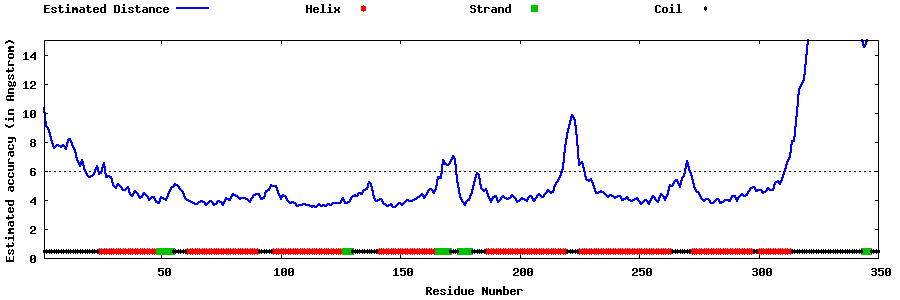 | |||
|
| |||
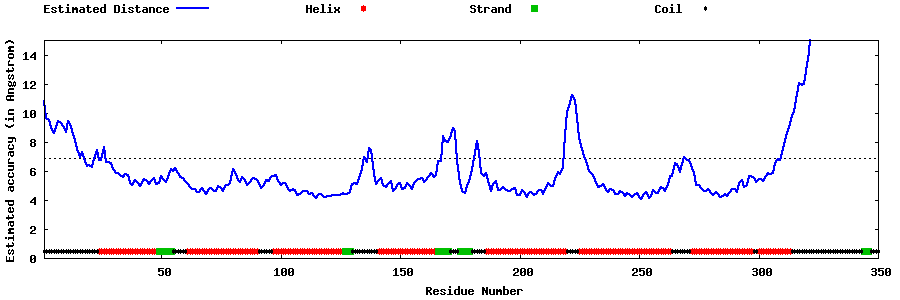 | |||
|
| |||
 | |||
|
| |||