

You can:
| Name | D(1A) dopamine receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | DRD1 |
| Synonym | D1 receptor D1A DADR Gpcr15 dopamine D1 receptor [ Show all ] |
| Disease | Unspecified Hypertension Pain Parkinson's disease Psychiatric disorder [ Show all ] |
| Length | 446 |
| Amino acid sequence | MRTLNTSAMDGTGLVVERDFSVRILTACFLSLLILSTLLGNTLVCAAVIRFRHLRSKVTNFFVISLAVSDLLVAVLVMPWKAVAEIAGFWPFGSFCNIWVAFDIMCSTASILNLCVISVDRYWAISSPFRYERKMTPKAAFILISVAWTLSVLISFIPVQLSWHKAKPTSPSDGNATSLAETIDNCDSSLSRTYAISSSVISFYIPVAIMIVTYTRIYRIAQKQIRRIAALERAAVHAKNCQTTTGNGKPVECSQPESSFKMSFKRETKVLKTLSVIMGVFVCCWLPFFILNCILPFCGSGETQPFCIDSNTFDVFVWFGWANSSLNPIIYAFNADFRKAFSTLLGCYRLCPATNNAIETVSINNNGAAMFSSHHEPRGSISKECNLVYLIPHAVGSSEDLKKEEAAGIARPLEKLSPALSVILDYDTDVSLEKIQPITQNGQHPT |
| UniProt | P21728 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | P21728 |
| 3D structure model | This predicted structure model is from GPCR-EXP P21728. |
| BioLiP | N/A |
| Therapeutic Target Database | T22118 |
| ChEMBL | CHEMBL2056 |
| IUPHAR | 214 |
| DrugBank | BE0000020 |
| Name | DIHYDREXIDINE |
|---|---|
| Molecular formula | C17H17NO2 |
| IUPAC name | (6aR,12bS)-5,6,6a,7,8,12b-hexahydrobenzo[a]phenanthridine-10,11-diol |
| Molecular weight | 267.328 |
| Hydrogen bond acceptor | 3 |
| Hydrogen bond donor | 3 |
| XlogP | 2.5 |
| Synonyms | (6aR,12bS)-10,11-Dihydroxy-5,6,6a,7,8,12b-hexahydro-benzo[a]phenanthridinium AC1O7G2G Lopac0_000380 UNII-Q3PJ4B4D0X (6aR,12bS)-5,6,6a,7,8,12b-hexahydrobenzo[a]phenanthridine-10,11-diol [ Show all ] |
| Inchi Key | BGOQGUHWXBGXJW-RHSMWYFYSA-N |
| Inchi ID | InChI=1S/C17H17NO2/c19-15-7-10-5-6-14-17(13(10)8-16(15)20)12-4-2-1-3-11(12)9-18-14/h1-4,7-8,14,17-20H,5-6,9H2/t14-,17-/m1/s1 |
| PubChem CID | 6603820 |
| ChEMBL | CHEMBL25856 |
| IUPHAR | N/A |
| BindingDB | 50010686 |
| DrugBank | N/A |
Structure | 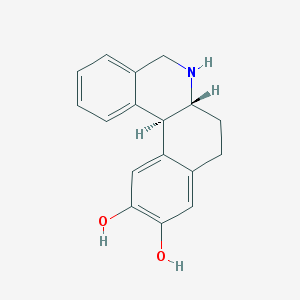 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| EC50 | 7.2 nM | PMID21862338 | BindingDB,ChEMBL |
| EC50 | 14.0 nM | PMID10360732, PMID7658429 | BindingDB,ChEMBL |
| EC50 | 19.0 nM | PMID17154515 | BindingDB,ChEMBL |
| IA | 121.0 % | PMID7658429 | ChEMBL |
| Intrinsic activity | 103.0 % | PMID21862338 | ChEMBL |
| Intrinsic activity | 110.0 nM | PMID17154515 | ChEMBL |
| Intrinsic activity | 121.0 % | PMID10360732 | ChEMBL |
| Ki | 6.2 nM | PMID22204903 | BindingDB,ChEMBL |
| Ki | 33.0 nM | PMID7658429, PMID10360732 | BindingDB,ChEMBL |
| Ki | 35.7 nM | PMID19744859 | BindingDB,ChEMBL |
| Ki | 1200.0 nM | PMID9784114 | BindingDB,ChEMBL |
| KL | 1200.0 nM | PMID8632409 | ChEMBL |
| Log 1/KL | -3.08 - | PMID8632409 | ChEMBL |
| nH | 0.65 - | PMID8558526 | ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417