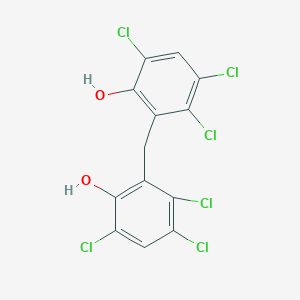
| Synonyms | MLS000028433
Surofene
NCGC00091195-02
TR-023301
NCGC00258902-01
ZINC1530968
NSC49115
Phenol, 2,2'-methylenebis[3,4,6-trichloro-
Hexascrub
Pre-Op
HMS2091J13
SBB058855
Isobac
Septofen
KBio3_001320
Spectrum4_000573
M0219
ST50880633
Dermadex
EINECS 200-733-8
Acigena (TN)
Exofene (TN)
Armohex
G-11
B 32
GERMA-MEDICA "MG"
Bis(2,3,5-tric hloro-6-hydroxyphenyl)methane
Hexachlorophane
Bis-2,3,5-trichlor-6-hydroxyfenylmethan
Hexachlorophene [USP:INN:BAN]
BRD-K99792991-001-02-6
CAS-70-30-4
2,2'-methanediylbis(3,4,6-trichlorophenol)
Compound G-11
2,3,3',5,5'-Hexachloro-6,6'-dihydroxydiphenylmethane
Steral
MolPort-001-785-865
Thera-Groom Pet Shampoo for Dogs for Veterinary Use Only
NCGC00091195-05
Turgex
NINDS_000630
Opera_ID_504
Hexachlorophenum [INN-Latin]
Phiso-Scrub
Hexosan
Rcra waste number U132
HMS501P12
SCHEMBL15579
KB-247348
Soy-dome
KSC-19-051
SR-01000721924
Methane, bis(2,3,5-trichloro-6-hydroxyphenyl)-
2,5,6-trichlorophenol)
DSSTox_CID_690
8054-98-6
Enditch Pet Shampoo
AKOS005449243
Fo stril
At-17
G-II
BDBM31712
H3P
Bis(2-hydroxy-3,6-trichlorophenyl)methane
Hexachlorophene (USP/INN)
Bis-2,5-trichlor-6-hydroxyfenylmethan
Hexachlorophene, United States Pharmacopeia (USP) Reference Standard
Brevity Blue Liquid Sanitizing Scouring Cream
139411-96-4
CCRIS 331
2,2'-Methylenebis(3,4,6-trichlorophenol)
CS-3866
MFCD00002171
Surgi-Cen
NC00512
Tox21_201350
NCGC00091195-08
UNII-IWW5FV6NK2
NSC-757055
Phenol, 2,2'-methylenebis(3,4,6-trichloro)-
Hexafen
Phisohex (TN)
Hilo Flea Powder
s4632
I14-19538
Septisol
KBio2_003915
Spectrum2_001105
KUC112427N
SR-01000721924-6
D0ZX2G
3,4,6-trichloro-2-[(2,3,5-trichloro-6-hydroxyphenyl)methyl]phenol
DTXSID6020690
ACGUYXCXAPNIKK-UHFFFAOYSA-N
Esaclorofene [DCIT]
AN-41934
Fostril (TN)
AT7 (TN)
Gamophene
Bilevon
Hexachlorofen
Bis(3,6-trichloro-2-hydroxyphenyl)methane
Hexachlorophene [INN]
Blockade Anti Bacterial Finish
C08039
2,2'-Dihydroxy-3,3',5,5',6,6'-hexachlorodiphenylmethane
CHEMBL496
2,2-Methylenebis(3,4,6-Trichlorophenol)
Staphene O
MLS001148404
Surofene (TN)
NCGC00091195-03
TRICHLOROPHENE
NCI-C02653
NSC757055
Hexachlorophenone
Phenol,2'-methylenebis[3,4,6-trichloro-
Hexide
PRE-OP II
HMS3259C19
SBI-0051403.P003
Isobac 20
SMR000058356
KBioGR_001006
Spectrum5_000792
MCULE-3028228047
Distodin
6,6'-Methylenebis(2,4,5-trichlorophenol)
Eleven
AI3-02372
Fascol
AS-10068
G-11 (TN)
B 32 (VAN)
GERMA-MEDICA (MG)
Bis(2,3,5-trichloro-6-hydroxyphenyl)methane
Hexachlorophen
Bis-2,3,5-trichloro-6-hydroxyfenylmethan
Hexachlorophene, PESTANAL(R), analytical standard
BRD-K99792991-001-18-2
0895AC
Caswell No. 566
2,2'-Methylene bis(3,4,6-trichlorophenol)
Cotofilm
2,4,6-trichlorophenol]
Steraskin
Nabac
Tox21_111099
NCGC00091195-06
UN 2875
NSC 49115
Pedigree Dog Shampoo Bar
Hexaclorofeno
Phisodan
Hexosan (TN)
REGID_for_CID_3598
HSDB 224
Scrubteam Surgical Spongebrush
KBio1_000630
SPBio_001210
KSC-285-117-1
SR-01000721924-2
Methane,3,5-trichloro-6-hydroxyphenyl)
3,3',5,5',6,6'-Hexachloro-2,2'-dihydroxydiphenylmethane
DSSTox_GSID_20690
AB00052010_17
EPA Pesticide Chemical Code 044901
Almederm
Fomac
AT-7
Gamophen
BIDD:ER0608
Hexa-Germ
Bis(3,5,6-trichloro-2-hydroxyphenyl)-methane
Hexachlorophene Solution, 5000 microg/mL in Dichloromethane
Bis[3,4,6-trichlorophenol], 2,2'-methylene-
BRN 2064407
2,2 -Methylene-bis-[3,4,6-trichlorophenol]
CHEBI:5693
2,2'-Methylenebis[3,4,6-trichlorophenol]
CTK9A1850
MLS-0072923.0001
Surgi-Cin
NCGC00091195-01
Tox21_302741
NCGC00256580-01
WLN: QR BG DG EG F1R BQ CG EG FG
NSC-9887
Phenol, 2,2'-methylenebis(3,4,6-trichloro-
Hexaphene-LV
Phisohex(TN)
HMS1920D07
SAM002554903
IDI1_000630
Septisol (TN)
KBio2_006483
Spectrum3_000450
LS-1597
ST24029562
DB00756
3,4,6-trichloro-2-[(3,5,6-trichloro-2-hydroxyphenyl)methyl]phenol
E-Z Scrub
Acigena
Exofene
API0002904
FT-0626955
B & b Flea Kontroller for Dogs Only
Germa-Medica
Bilevon (TN)
Hexachlorofen [Czech]
Bis-(2-hydroxy-3,5,6-trichlorophenyl)methane
Hexachlorophene [UN2875] [Poison]
BP-30177
C13H6Cl6O2
2,2'-Dihydroxy-3,5,6,3',5',6'-hexachlorodiphenylmethane
cid_3598
2,3',5,5',6,6'-hexachlorodiphenylmethane
Ster-zac
MLS002152906
Tersaseptic
NCGC00091195-04
Trisophen
Neosept V
NSC9887
Hexachlorophenum
Phenol,2'-methylenebis[3,5,6-trichloro-
Hexophene
RCRA waste no. U132
HMS3715P21
SC-51022
IWW5FV6NK2
Solu-Heks
KBioSS_001347
Spectrum_000867
Methane, bis(2,3,5-trichloro-6-hydroxyphenyl)
DivK1c_000630
70-30-4
En-Viron D Concentrated Phenolic Disinfectant
AJ-26716
Fesia-sin
AT 7
G-Eleven
B32
Germa-Medica (TN)
Bis(2-hydroxy-3,5,6-trichlorophenyl)methane
Bis-2,3,5-trichloro-6-hydroxyfenylmethan [Czech]
Hexachlorophene, Pharma
Brevity Blue Liquid Bacteriostatic Scouring Cream
1195388-85-2
CCG-39768
2,2'-Methylene-bis-[3,4,6-trichlorophenol]
CPD000058356
2,5,6,3',5',6'-hexachlorodiphenylmethane
Methylenebis(3,4,6-trichlorophenol)
STK377478
Nabac 25 ec
Tox21_111099_1
NCGC00091195-07
UN2875
NSC-49115
Pharmakon1600-01500328
Hexaclorofeno [INN-Spanish]
pHisoHex
Hilo Cat Flea Powder
Ritosept
HY-12637
Septi-Soft
KBio2_001347
SPECTRUM1500328
KUC106447N
SR-01000721924-5
3,4,6-trichloro-2-[(2,3,5-trichloro-6-hydroxy-phenyl)methyl]phenol
DSSTox_RID_75738
AC1L1GAE
Esaclorofene
Almederm (TN)
Fostril
AT17 (TN)
Gamophen (TN)
BIDD:GT0722
Hexabalm
Bis(3,5,6-trichloro-2-hydroxyphenyl)methane
Hexachlorophene Solution, 5000 microg/mL in Methanol
Bivelon
BSPBio_002100
2,2',3,3',5,5'-Hexachloro-6,6'-dihydroxydiphenylmethane
2,2-Methylene-bis-[3,4,6-trichlorophenol]
D00859 [ Show all ] |
|---|


![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417