

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 | |
| | | | | | | | | | | | | | | | | | | | | |
| MGPIGAEADENQTVEEMKVEQYGPQTTPRGELVPDPEPELIDSTKLIEVQVVLILAYCSIILLGVIGNSLVIHVVIKFKSMRTVTNFFIANLAVADLLVNTLCLPFTLTYTLMGEWKMGPVLCHLVPYAQGLAVQVSTITLTVIALDRHRCIVYHLESKISKRISFLIIGLAWGISALLASPLAIFREYSLIEIIPDFEIVACTEKWPGEEKSIYGTVYSLSSLLILYVLPLGIISFSYTRIWSKLKNHVSPGAANDHYHQRRQKTTKMLVCVVVVFAVSWLPLHAFQLAVDIDSQVLDLKEYKLIFTVFHIIAMCSTFANPLLYGWMNSNYRKAFLSAFRCEQRLDAIHSEVSVTFKAKKNLEVRKNSGPNDSFTEATNV | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSSCCCCCSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 999988888776740125455798888788788987643456444405999999999999999989998872652147999848789999999999999999999999999828778926789879999999999999999999997098976448654247788884179999999999999999244773221789727996038860266999999999999999999999999999999998056899840368999741499999999999999979999999999975664551479999999999999999999999996899999999998188787887787778775656761100179999998878989 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 | |
| | | | | | | | | | | | | | | | | | | | | |
| MGPIGAEADENQTVEEMKVEQYGPQTTPRGELVPDPEPELIDSTKLIEVQVVLILAYCSIILLGVIGNSLVIHVVIKFKSMRTVTNFFIANLAVADLLVNTLCLPFTLTYTLMGEWKMGPVLCHLVPYAQGLAVQVSTITLTVIALDRHRCIVYHLESKISKRISFLIIGLAWGISALLASPLAIFREYSLIEIIPDFEIVACTEKWPGEEKSIYGTVYSLSSLLILYVLPLGIISFSYTRIWSKLKNHVSPGAANDHYHQRRQKTTKMLVCVVVVFAVSWLPLHAFQLAVDIDSQVLDLKEYKLIFTVFHIIAMCSTFANPLLYGWMNSNYRKAFLSAFRCEQRLDAIHSEVSVTFKAKKNLEVRKNSGPNDSFTEATNV | |
| 723453433222122323243233322332313333333233222210010000110120010023122100000001340210001002000200100000001010000016330103001100000100000000000000000002000000233223310000000001000000100000120231423543210000020134434202100000102211231010002000100010233333454454334322000000000000000002230000001002232242320100000000000100000000000014300400230020123344344533443344443434444344433344154 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 | | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSSCCCCCSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MGPIGAEADENQTVEEMKVEQYGPQTTPRGELVPDPEPELIDSTKLIEVQVVLILAYCSIILLGVIGNSLVIHVVIKFKSMRTVTNFFIANLAVADLLVNTLCLPFTLTYTLMGEWKMGPVLCHLVPYAQGLAVQVSTITLTVIALDRHRCIVYHLESKISKRISFLIIGLAWGISALLASPLAIFREYSLIEIIPDFEIVACTEKWPGEEKSIYGTVYSLSSLLILYVLPLGIISFSYTRIWSKLKNHVSPGAANDHYHQRRQKTTKMLVCVVVVFAVSWLPLHAFQLAVDIDSQVLDLKEYKLIFTVFHIIAMCSTFANPLLYGWMNSNYRKAFLSAFRCEQRLDAIHSEVSVTFKAKKNLEVRKNSGPNDSFTEATNV | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.21 | 0.23 | 0.78 | 3.08 | Download | -------------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLM--TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-----GAVVCMLQFPSP-SWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS-KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------------ | |||||||||||||||||||
| 2 | 4n6hA | 0.23 | 0.23 | 0.75 | 4.00 | Download | ------------------------------------------------LAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALA-TSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTRPR-----DGAVVCMLQFPSP-SWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSG-SKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKP-------------------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.22 | 0.23 | 0.79 | 3.83 | Download | -------------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATST-LPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGA-----VVCMLQFPSP-SWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS-KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------------ | |||||||||||||||||||
| 4 | 4djh | 0.25 | 0.24 | 0.75 | 1.54 | Download | ---------------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPLKAKIINICIWLLSSSVGISAIVLGGTKVR---EDVDVIECSLQFPDDDYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP--------------------------------------- | |||||||||||||||||||
| 5 | 4djh | 0.25 | 0.24 | 0.75 | 1.17 | Download | ---------------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVT-TTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRE---DVDVIECSLQFPDDDYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAA------LSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP--------------------------------------- | |||||||||||||||||||
| 6 | 4n6hA | 0.23 | 0.23 | 0.78 | 3.29 | Download | ---------------------------------------GARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALA-TSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-----GAVVCMLQFP-SPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSG-SKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------------ | |||||||||||||||||||
| 7 | 4djh | 0.26 | 0.24 | 0.74 | 1.71 | Download | ------------------------------------------------IPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDTPLKAKIINICIWLLSSSVGISAIVLGGTKVR---EDVDVIECSLQFPDDDYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGAYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCF---------------------------------------- | |||||||||||||||||||
| 8 | 4ea3A | 0.25 | 0.21 | 0.73 | 4.81 | Download | ---------------------------------------------PLGLKVTIVGLYLAVCVGGLLGNCLVMYVILRHTKMKTATNIYIFNLALADTLVLL-TLPFQGTDILLGFWPFGNALCKTVIAIDYYNMFTSTFTLTAMSVDRYVAICHPT-----SSKAQAVNVAIWALASVVGVPVAIM---GSAQVEDEEIECLVEIPTP---QDYWGPVFAICIFLFSFIVPVLVISVCYSLMIRRLRGVRLLSGS-REKDRNLRRITRLVLVVVAVFVGCWTPVQVFVLAQGLGVQPSS-ETAVAILRFCTALGYVNSCLNPILYAFLDENFKACFR-------------------------------------------- | |||||||||||||||||||
| 9 | 4n6hA | 0.21 | 0.23 | 0.78 | 3.17 | Download | -------------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLM--TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-----GAVVCMLQFPSP-SWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS-KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------------ | |||||||||||||||||||
| 10 | 5c1mA | 0.22 | 0.19 | 0.77 | 3.73 | Download | ----------------------------------GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVRYTKMKTATNIYIFNLALADALAT-STLPFQSVNYLMGTWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALRTPRNAKIVNVCNWILSSAIGLPVMFMATTKYR-----QGSIDCTLTFSHP-TWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLK-SVRMLSGSKEKDRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKALI-TIPETTFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF--------------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
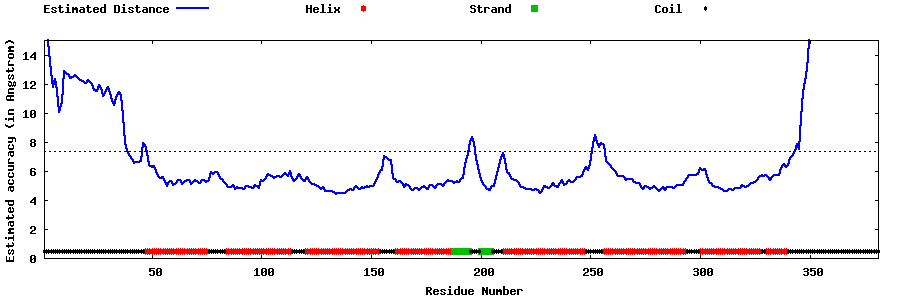
| Generated 3D models | Estimated local accuracy of models | ||
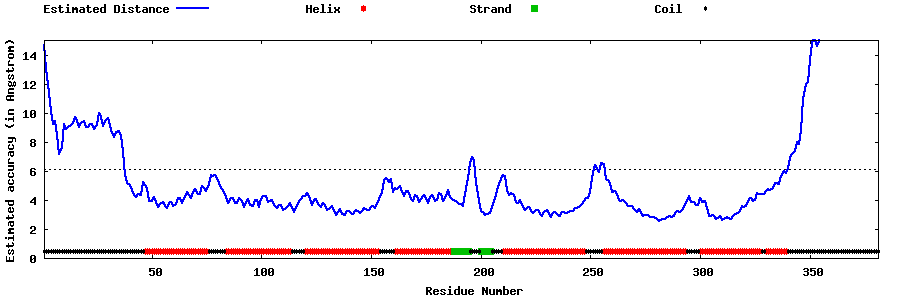 | |||
|
|
|||
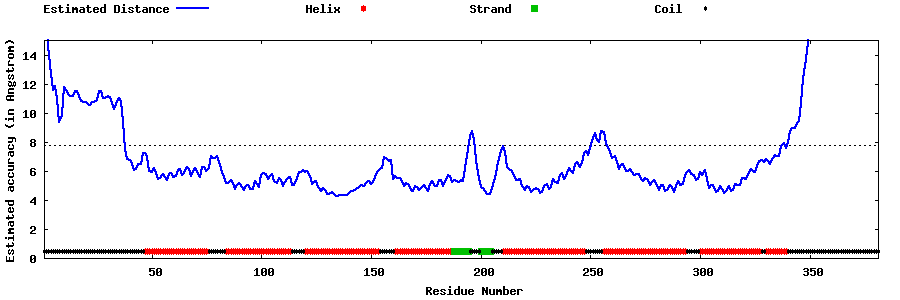 | |||
|
| |||
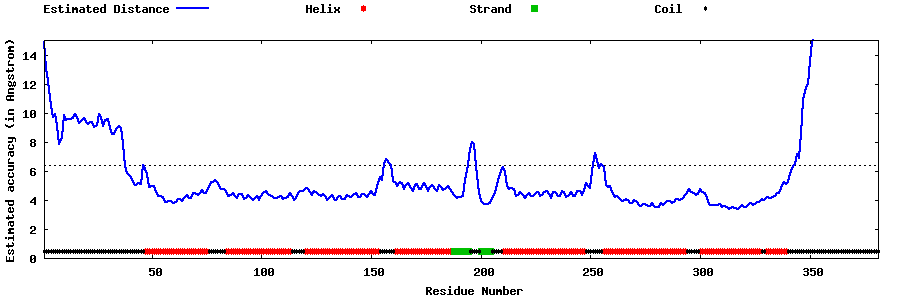 | |||
|
| |||
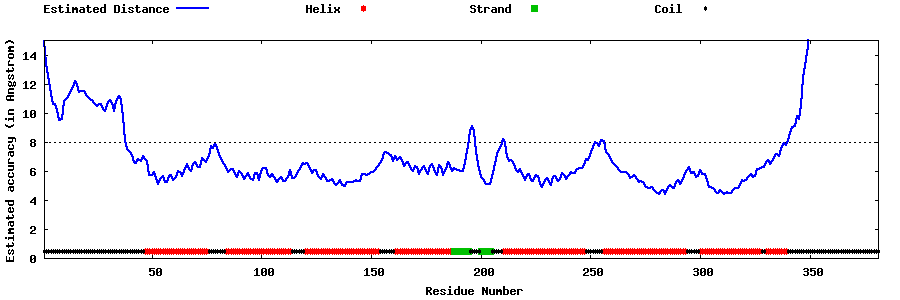 | |||
|
| |||
 | |||
|
| |||