

You can:
| Name | 5-hydroxytryptamine receptor 1A |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | HTR1A |
| Synonym | 5-HT-1A 5-HT1A serotonin receptor 1A 5-HT1A receptor 5-hydroxytryptamine (serotonin) receptor 1A, G protein-coupled [ Show all ] |
| Disease | Urinary incontinence Generalized anxiety disorder Generalized anxiety disorder; Social phobia Hypertension Hypoactive sexual desire disorder [ Show all ] |
| Length | 422 |
| Amino acid sequence | MDVLSPGQGNNTTSPPAPFETGGNTTGISDVTVSYQVITSLLLGTLIFCAVLGNACVVAAIALERSLQNVANYLIGSLAVTDLMVSVLVLPMAALYQVLNKWTLGQVTCDLFIALDVLCCTSSILHLCAIALDRYWAITDPIDYVNKRTPRRAAALISLTWLIGFLISIPPMLGWRTPEDRSDPDACTISKDHGYTIYSTFGAFYIPLLLMLVLYGRIFRAARFRIRKTVKKVEKTGADTRHGASPAPQPKKSVNGESGSRNWRLGVESKAGGALCANGAVRQGDDGAALEVIEVHRVGNSKEHLPLPSEAGPTPCAPASFERKNERNAEAKRKMALARERKTVKTLGIIMGTFILCWLPFFIVALVLPFCESSCHMPTLLGAIINWLGYSNSLLNPVIYAYFNKDFQNAFKKIIKCKFCRQ |
| UniProt | P08908 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | P08908 |
| 3D structure model | This predicted structure model is from GPCR-EXP P08908. |
| BioLiP | N/A |
| Therapeutic Target Database | T78709 |
| ChEMBL | CHEMBL214 |
| IUPHAR | 1 |
| DrugBank | BE0000291 |
| Name | 8-OH-Dpat |
|---|---|
| Molecular formula | C16H25NO |
| IUPAC name | 7-(dipropylamino)-5,6,7,8-tetrahydronaphthalen-1-ol |
| Molecular weight | 247.382 |
| Hydrogen bond acceptor | 2 |
| Hydrogen bond donor | 1 |
| XlogP | 4.1 |
| Synonyms | AC1Q7AL7 BRD-A48015106-004-03-0 D0O3GF LS-94994 NCGC00024986-02 [ Show all ] |
| Inchi Key | ASXGJMSKWNBENU-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C16H25NO/c1-3-10-17(11-4-2)14-9-8-13-6-5-7-16(18)15(13)12-14/h5-7,14,18H,3-4,8-12H2,1-2H3 |
| PubChem CID | 1220 |
| ChEMBL | CHEMBL56 |
| IUPHAR | 31, 7 |
| BindingDB | 21393 |
| DrugBank | N/A |
Structure | 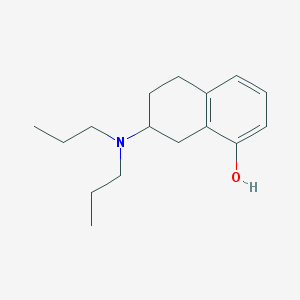 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| %max | 100.0 - | PMID18817363 | ChEMBL |
| EC50 | 2.6 nM | PMID15887953 | ChEMBL |
| EC50 | 5.82 nM | PMID11101361 | BindingDB,ChEMBL |
| EC50 | 8.0 nM | PMID24050112 | ChEMBL |
| EC50 | 10.9 nM | PMID15887953 | BindingDB,ChEMBL |
| EC50 | 12.0 nM | PMID26081758 | BindingDB |
| EC50 | 12.02 nM | PMID26081758 | ChEMBL |
| EC50 | 15.0 nM | PMID23279866 | BindingDB,ChEMBL |
| EC50 | 26.0 nM | PMID18834188 | BindingDB,ChEMBL |
| EC50 | 28.18 nM | PMID17300168 | ChEMBL |
| EC50 | 32.36 nM | PMID17803293 | ChEMBL |
| EC50 | 51.0 nM | PMID26081758 | BindingDB |
| EC50 | 51.29 nM | PMID26081758 | ChEMBL |
| EC50 | 76.0 nM | PMID11448222 | ChEMBL |
| Effect | 100.0 % | PMID11448222 | ChEMBL |
| Emax | 26.0 % | PMID20605276 | ChEMBL |
| Emax | 35.9 % | PMID17803293 | ChEMBL |
| Emax | 44.5 % | PMID26081758 | ChEMBL |
| Emax | 82.0 % | PMID17300168 | ChEMBL |
| Emax | 84.8 % | PMID26081758 | ChEMBL |
| Emax | 100.0 % | PMID20185311, PMID22145629, PMID27689727, PMID23252794, PMID24900506, PMID11728188, PMID24900763, PMID15887953, PMID18834188 | ChEMBL |
| IC50 | 0.33 nM | PMID23466604 | ChEMBL |
| IC50 | 0.51 nM | PMID23582449 | ChEMBL |
| IC50 | 0.53 nM | PMID26988801, PMID27876250 | ChEMBL |
| IC50 | 0.59 nM | PMID18588282 | ChEMBL |
| IC50 | 1.1 nM | PMID18983139 | ChEMBL |
| IC50 | 5.3 nM | PMID26988801, PMID27876250 | BindingDB |
| IC50 | 6.0 nM | PMID24262884 | ChEMBL |
| Intrinsic activity | 0.9 - | PMID18433113 | ChEMBL |
| Kd | 0.088 - | PMID11101361 | ChEMBL |
| Kd | 0.398 - 1000.0 nM | PMID10431754, PMID9550290, PMID15628665, PMID11101361 | IUPHAR |
| Ki | 0.06 nM | PMID7984267 | PDSP,BindingDB |
| Ki | 0.17 nM | PMID21726069 | BindingDB,ChEMBL |
| Ki | 0.21 nM | PMID23466604 | ChEMBL |
| Ki | 0.36 nM | PMID11132243 | PDSP |
| Ki | 0.398 - 3.98 nM | PMID1386736, PMID9760039, PMID9550290, PMID15628665, PMID9495870, PMID10611634, PMID12527336, PMID9067310 | IUPHAR |
| Ki | 0.4 nM | PMID25557493, Bioorg. Med. Chem. Lett., (1997) 7:21:2759, PMID24805037 | ChEMBL |
| Ki | 0.4 nM | , PMID25557493, PMID24805037 | BindingDB |
| Ki | 0.47 nM | PMID9067310 | PDSP,BindingDB |
| Ki | 0.5 nM | PMID8340922 | BindingDB,ChEMBL |
| Ki | 0.58 nM | PMID9067310 | PDSP,BindingDB |
| Ki | 0.64 nM | PMID9067310 | PDSP,BindingDB |
| Ki | 0.66 nM | PMID18983139 | ChEMBL |
| Ki | 0.7 nM | Hoyer et al., PMID1986 | PDSP |
| Ki | 0.71 nM | PMID25695425 | ChEMBL |
| Ki | 0.71 nM | PMID25695425 | BindingDB |
| Ki | 0.7943 nM | PMID18433113 | ChEMBL |
| Ki | 0.794328 nM | PMID18433113 | BindingDB |
| Ki | 0.81 nM | PMID7984267 | PDSP,BindingDB |
| Ki | 0.83 nM | Hoyer et al., PMID1986, PMID11277522 | PDSP,ChEMBL |
| Ki | 0.83 nM | PMID11277522 | BindingDB |
| Ki | 1.0 nM | PMID2078271 | BindingDB |
| Ki | 1.2 nM | PMID20041669 | BindingDB,ChEMBL |
| Ki | 1.25 nM | PMID7984267 | PDSP,BindingDB |
| Ki | 1.259 nM | PMID21469694 | ChEMBL |
| Ki | 1.49 nM | PMID9871775 | ChEMBL |
| Ki | 1.5 nM | PMID9871775 | BindingDB |
| Ki | 1.6 nM | PMID8097537, PMID8459396, PMID8097538 | BindingDB,ChEMBL |
| Ki | 1.9 nM | PMID7984267, PMID9083484 | PDSP,BindingDB,ChEMBL |
| Ki | 2.1 nM | PMID17649988, PMID18800769 | PDSP |
| Ki | 2.1 nM | PMID17649988, PMID18800769 | BindingDB,ChEMBL |
| Ki | 2.65 nM | PMID28063784, PMID25759032 | ChEMBL |
| Ki | 2.7 nM | PMID28063784, PMID25759032 | BindingDB |
| Ki | 3.0 nM | PMID18834188, PMID15887953 | PDSP,BindingDB,ChEMBL |
| Ki | 3.16 nM | PMID10611634 | PDSP,BindingDB |
| Ki | 3.38 nM | PMID7984267 | PDSP,BindingDB |
| Ki | 3.388 nM | PMID18817363, PMID24900763, PMID23252794, PMID24900506 | ChEMBL |
| Ki | 3.39 nM | PMID18817363 | BindingDB |
| Ki | 3.4 nM | PMID23252794, PMID24900506 | BindingDB |
| Ki | 3.44 nM | PMID11728188, PMID11448222 | BindingDB,ChEMBL |
| Ki | 3.467 nM | PMID10514291 | ChEMBL |
| Ki | 3.715 nM | PMID22145629, PMID20185311 | ChEMBL |
| Ki | 3.72 nM | PMID22145629, PMID20185311 | BindingDB |
| Ki | 3.76 nM | PMID8461029 | PDSP,BindingDB |
| Ki | 3.8 nM | PMID7984267, PMID15055991 | PDSP,BindingDB,ChEMBL |
| Ki | 4.57 nM | PMID7984267 | PDSP,BindingDB |
| Ki | 5.1 nM | PMID18269229 | BindingDB,ChEMBL |
| Ki | 6.9 nM | PMID9686407 | PDSP,BindingDB |
| Ki | 26.91 nM | PMID7984267 | PDSP,BindingDB |
| Ki | 27.1 nM | PMID8461029 | PDSP,BindingDB |
| Ki | 78.0 nM | PMID8155646 | PDSP,BindingDB |
| Ki | 3300.0 nM | PMID19053888 | PDSP,BindingDB,ChEMBL |
| Ki | 3981.0 nM | PMID18433113 | BindingDB |
| Ki | 3981.07 nM | PMID18433113 | ChEMBL |
| Log Ki | 8.47 nM | PMID10425105 | ChEMBL |
| Max | 100.0 % | PMID10514291, PMID10425105 | ChEMBL |
| Others | 134.0 % | PMID8941384 | ChEMBL |
| pD2 | 7.6 - | PMID18817363, PMID10514291, PMID23252794, PMID11728188, PMID10425105, PMID24900763, PMID24900506 | ChEMBL |
| pD2 | 7.83 - | PMID22145629, PMID20185311 | ChEMBL |
| pD2 | 8.49 - | PMID27689727 | ChEMBL |
| pD2 | 9.27 - | PMID20605276 | ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417