

You can:
| Name | Histamine H4 receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | HRH4 |
| Synonym | Pfi-013 SP9144 HH4R H4R H4 receptor [ Show all ] |
| Disease | Allergic rhinitis Asthma Inflammatory disease Rheumatoid arthritis |
| Length | 390 |
| Amino acid sequence | MPDTNSTINLSLSTRVTLAFFMSLVAFAIMLGNALVILAFVVDKNLRHRSSYFFLNLAISDFFVGVISIPLYIPHTLFEWDFGKEICVFWLTTDYLLCTASVYNIVLISYDRYLSVSNAVSYRTQHTGVLKIVTLMVAVWVLAFLVNGPMILVSESWKDEGSECEPGFFSEWYILAITSFLEFVIPVILVAYFNMNIYWSLWKRDHLSRCQSHPGLTAVSSNICGHSFRGRLSSRRSLSASTEVPASFHSERQRRKSSLMFSSRTKMNSNTIASKMGSFSQSDSVALHQREHVELLRARRLAKSLAILLGVFAVCWAPYSLFTIVLSFYSSATGPKSVWYRIAFWLQWFNSFVNPLLYPLCHKRFQKAFLKIFCIKKQPLPSQHSRSVSS |
| UniProt | Q9H3N8 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | Q9H3N8 |
| 3D structure model | This predicted structure model is from GPCR-EXP Q9H3N8. |
| BioLiP | N/A |
| Therapeutic Target Database | T26500 |
| ChEMBL | CHEMBL3759 |
| IUPHAR | 265 |
| DrugBank | BE0000146 |
| Name | JNJ-7777120 |
|---|---|
| Molecular formula | C14H16ClN3O |
| IUPAC name | (5-chloro-1H-indol-2-yl)-(4-methylpiperazin-1-yl)methanone |
| Molecular weight | 277.752 |
| Hydrogen bond acceptor | 2 |
| Hydrogen bond donor | 1 |
| XlogP | 2.3 |
| Synonyms | 5-chloro-2-[(4-methylpiperazin-1-yl)carbonyl]-1h-indole FT-0747430 MolPort-002-651-947 SCHEMBL186434 1-[(5-CHLORO-1H-INDOL-2-YL)CARBONYL]-4-METHYL-PIPERAZINE [ Show all ] |
| Inchi Key | HUQJRYMLJBBEDO-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C14H16ClN3O/c1-17-4-6-18(7-5-17)14(19)13-9-10-8-11(15)2-3-12(10)16-13/h2-3,8-9,16H,4-7H2,1H3 |
| PubChem CID | 4908365 |
| ChEMBL | CHEMBL129198 |
| IUPHAR | 1279, 1278 |
| BindingDB | 22566 |
| DrugBank | N/A |
Structure | 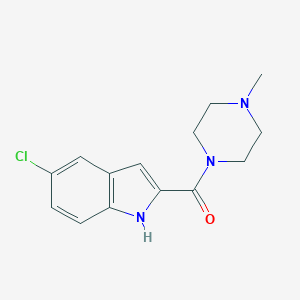 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| EC50 | 15.85 nM | PMID19773175 | ChEMBL |
| EC50 | 25.0 nM | PMID24697360 | BindingDB |
| EC50 | 25.12 nM | PMID24697360 | ChEMBL |
| Emax | -58.7 % | PMID19773175 | ChEMBL |
| Emax | -0.74 % | PMID21944853 | ChEMBL |
| IC50 | 5.3 nM | PMID21920751 | BindingDB,ChEMBL |
| IC50 | 30.0 nM | PMID21920751 | BindingDB,ChEMBL |
| IC50 | 40.0 nM | PMID16366610 | ChEMBL |
| IC50 | 86.0 nM | PMID16366610, PMID22749391 | BindingDB,ChEMBL |
| IC50 | 199.0 nM | PMID21920751 | BindingDB,ChEMBL |
| IC50 | 199.53 nM | PMID24697360 | ChEMBL |
| IC50 | 200.0 nM | PMID24697360 | BindingDB |
| IC50 | 221.4 nM | PMID21920751 | BindingDB,ChEMBL |
| Intrinsic activity | -0.54 - | PMID27007611 | ChEMBL |
| Kb | 12.0 nM | PMID19791743 | ChEMBL |
| Kb | 19.1 nM | PMID27007611 | ChEMBL |
| Kb | 29.0 nM | PMID27007611 | ChEMBL |
| Kd | 3.6 nM | PMID14722321 | IUPHAR |
| Kd | 3.981 nM | PMID23558237 | ChEMBL |
| Kd | 7.244 nM | PMID12954048 | ChEMBL |
| Kd | 7.943 nM | PMID16366610 | ChEMBL |
| Kd | 10.0 nM | PMID15454206 | BindingDB,ChEMBL |
| Ki | 3.311 nM | PMID21920751 | ChEMBL |
| Ki | 4.0 nM | Med Chem Res, (2004) 13:8:619, PMID12954048, PMID22189138, PMID16366610, PMID22153663, PMID21458260 | BindingDB,ChEMBL |
| Ki | 4.1 nM | PMID14722321 | BindingDB |
| Ki | 4.169 nM | PMID19053770 | ChEMBL |
| Ki | 4.17 nM | PMID19053770 | BindingDB |
| Ki | 4.5 nM | PMID25993395 | BindingDB,ChEMBL |
| Ki | 4.898 nM | PMID26718844 | ChEMBL |
| Ki | 4.9 nM | PMID26718844 | BindingDB |
| Ki | 5.0 nM | PMID25455490, PMID24495018, PMID25595684 | ChEMBL |
| Ki | 5.0 nM | PMID25455490, PMID24495018, PMID25595684 | BindingDB |
| Ki | 5.01 - 15.8 nM | PMID15947036, PMID14722321, PMID16854056 | IUPHAR |
| Ki | 6.0 nM | PMID22153663 | BindingDB,ChEMBL |
| Ki | 6.31 nM | PMID23668417, PMID22749391 | BindingDB,ChEMBL |
| Ki | 6.8 nM | PMID21955944, PMID22189138, PMID21920751 | BindingDB,ChEMBL |
| Ki | 8.0 nM | PMID21955944, PMID22189138, PMID21920751 | BindingDB,ChEMBL |
| Ki | 12.0 nM | PMID18983139, PMID18817367, PMID18811133 | BindingDB,ChEMBL |
| Ki | 12.02 nM | PMID18983139, PMID18817367, PMID18811133 | ChEMBL |
| Ki | 13.0 nM | PMID27007611 | BindingDB,ChEMBL |
| Ki | 14.0 nM | PMID19773175 | BindingDB,ChEMBL |
| Ki | 15.85 nM | PMID18357976, PMID16854056, PMID22003888 | BindingDB,ChEMBL |
| Ki | 16.0 nM | PMID15947036 | BindingDB |
| Ki | 17.0 nM | PMID18459760, Med Chem Res, (2004) 13:8:619 | BindingDB,ChEMBL |
| Ki | 18.62 nM | PMID21944853 | BindingDB,ChEMBL |
| Ki | 19.95 nM | MedChemComm, (2013) 4:1:193, PMID15454206 | ChEMBL |
| Ki | 35.0 nM | PMID26718844 | BindingDB |
| Ki | 35.48 nM | PMID26718844 | ChEMBL |
| Ki | 38.0 nM | PMID20299215 | BindingDB |
| Ki | 46.0 nM | PMID22189138 | BindingDB,ChEMBL |
| Ki | 50.12 nM | PMID19773175 | ChEMBL |
| Ki | 69.0 nM | PMID27007611 | BindingDB,ChEMBL |
| Ki | 118.0 nM | PMID22189138 | BindingDB,ChEMBL |
| Ki | 3999450.0 nM | Med Chem Res, (2004) 13:8:619 | ChEMBL |
| Ki | 16982400.0 nM | Med Chem Res, (2004) 13:8:619 | ChEMBL |
| Kinact | 16.0 nM | PMID19791743 | ChEMBL |
| pKb | 8.31 - | PMID18983139 | ChEMBL |
| pKb | 8.37 - | PMID18817367, PMID18811133 | ChEMBL |
| T1/2 | 1.033 hr | PMID22153663 | ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417