

Go to page (83 pages in total): [First page] << < 24 25 26 27 28 29 30 31 32 33 34 > >> [Last page] [All entries in one page].
| # | Pfam ID | D-I-TASSER models ranked by estimated TM-score (click to view) |
Full name and representative sequence |
| 2801 |
PF04923 (Ninjurin) |
 Estimated TM-score=0.71; hard target. |
>Ninjurin (Length=102) INVYQQKKSLAQGMMDLALLSANANQLRYVLESCQNHPFFYVNLVFVSTSIFVQVVVGFGLIWKSQYNMNNPDDYRAATR INNLVSIGIFIITLVNVMLSAF |
| 2802 |
PF13396 (PLDc_N) |
 Estimated TM-score=0.71; hard target. |
>Phospholipase_D-nuclease N-terminal (Length=41) ILWVVAIFDCAKSDHPNKIGWIVVIILLPFLGSILYFIFGR |
| 2803 |
PF04659 (Arch_fla_DE) |
 Estimated TM-score=0.71; hard target. |
>Archaeal flagella protein (Length=93) KARLEKLPEDVVSTMIALKWLGFLIDRVGIQNLERVLEFYYEIGWISEEVLNQLLRYAKGTRPHHRDPEWKPADKLTVQD HLISLLFIERLRG |
| 2804 |
PF15923 (DUF4745) |
 Estimated TM-score=0.71; easy target. |
>Domain of unknown function (DUF4745) (Length=128) AGGCHHAMPHSTPIADIQQGISKYLDALNVFCRASTFLTDLFSTVFRNSHYSKAAMQLKDVQEHVMEAASRLTSAIKPEI AKMLMELSAGAANFTDQKEFSLQDIEVLGRCFLTVVQVHFQFLTQALQ |
| 2805 |
PF07583 (PSCyt2) |
 Estimated TM-score=0.71; very hard target. |
>Protein of unknown function (DUF1549) (Length=207) PIDAFILHRLEAQQLKPSPEADRTTLIRRLSLDLTGLPPSVKQVQEFLADTKPGAYERLVDRLLASPHYGERWARHWLDV ARYADSNGFTIDGPRSIWKYRDWVIEAINANMPFDQFVTEQLAGDLLPKPTTEQLIATGFHRNTLINQEGGTNPEQFRVE AVVDRVNTTGAAFLGLTVGCAQCHKHKYDPITQRDFYQLYAIFNSTA |
| 2806 |
PF08318 (COG4) |
 Estimated TM-score=0.71; hard target. |
>COG4 transport protein (Length=322) LQQATMQLRHLITQKFDESVKKDDLASVERFFKIFPLLGMHDHGIEKFSQYICHKLNAKAQKELRTSMDTAKAEKRTVVA FADTLTILLENIARVVEVNQPIIETYYGKGRLHQMVRVLQEECDQEVKICLLEFNKGRHIQRRVAQINEYIKTGGSGGSS VAGHYRKPSGGSMDKLNPKDIDSLIGEITIMHSRAELYVKFIKRRVARDLDSSTLDEEKKRTALAGVEKCLKDSDLSRQM QELLGIYLLFERFFMEESVLKAIALDAHEPGQQCSSMIDDVFFIVRKCIRRANTTQSLDGICAVINNAATELENDFIAAI KE |
| 2807 |
PF03684 (UPF0179) |
 Estimated TM-score=0.71; very hard target. |
>Uncharacterised protein family (UPF0179) (Length=141) TITLIGVRLAKVGTEFIFKGPAPECEDCRFKNSCMNLNPGRKYRIVNIRTTTTHSCDVHDEGVQAVEIVESPIISAIESK KAFKGSKIVFEPPLCKRSECNMHDLCHPIGLEFGDKCTISDVIGDAPDECMKGLSLKVVEL |
| 2808 |
PF15604 (Ntox15) |
 Estimated TM-score=0.71; very hard target. |
>Novel toxin 15 (Length=146) QHPAEFARQLSRQYEALSYLTVDEYLKGRNLYNTQKRAPTPPSLELQREDLEKKIVNRLVDDYRFDPDKAEKKAVEAVQA LAALHEPDQSLQGTHQLAAGEIVWGNSSVNSSIGGQHKAGLARLDEKARDAQVAGKGHRYLNVRVV |
| 2809 |
PF16459 (Phage_TAC_13) |
 Estimated TM-score=0.71; easy target. |
>Phage tail assembly chaperone, TAC (Length=104) DSLKEMGAFTGAPVEKEITWKQGKKELTATVYVRRLSYRTAVSDIQAMKNNSDAVAGRIAASICDEKGKPIFTPEDITGD ADPDRGPLDGNLTMALLSAIGEVN |
| 2810 |
PF11324 (DUF3126) |
 Estimated TM-score=0.71; hard target. |
>Protein of unknown function (DUF3126) (Length=59) ELQKLQGFLRQSFGNQGIKVTQARKNPDDADVLLGERQIGEIMVDDEDGDRSFAFAMKV |
| 2811 |
PF02534 (T4SS-DNA_transf) |
 Estimated TM-score=0.71; trivial target. |
>Type IV secretory system Conjugative DNA transfer (Length=502) RWADKKDIQAAGLLPRPRTVVELVSGKHPPTSSGVYVGGWQDKDGKFHYLRHNGPEHVLTYAPTRSGKGVGLVVPTLLSW AHSAVITDLKGELWALTAGWRKKHARNKVVRFEPASAQGSACWNPLDEIRLGTEYEVGDVQNLATLIVDPDGKGLESHWQ KTSQALLVGVILHALYKAKNEGTPATLPSVDGMLADPNRDVGELWMEMTTYGHVDGQNHPAVGSAARDMMDRPEEESGSV LSTAKSYLALYRDPVVARNVSKSDFRIKQLMHHDDPVSLFIVTQPNDKARLRPLVRVMVNMIVRLLADKMDFENGRPVAH YKHRLLMMLDEFPSLGKLEILQESLAFVAGYGIKCYLICQDINQLKSRETGYGHDESITSNCHVQNAYPPNRVETAEHLS KLTGTTTIVKEQITTSGRRTSALLGNVSRTFQEVQRPLLTPDECLRMPGPKKSADGSIEEAGDMVVYVAGYPAIYGKQPL YFKDPIFQARAAVPAPKVSDKL |
| 2812 |
PF17323 (ToxS) |
 Estimated TM-score=0.71; very hard target. |
>Trans-membrane regulatory protein ToxS (Length=149) YWGSDLKVEQVLTAKEWQSKMVTVITDSLQEESVGPLRKVDVTSNVKYLPNGSYIRVATVRLYANNAESENVINISETGE WDISDNYLLVSPIEFKDVSSAQSKDFTADQLKLITQIFKMDAQQSRRIDIVNEKTLLLTSLSHGSTVLF |
| 2813 |
PF16429 (DUF5026) |
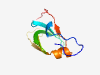 Estimated TM-score=0.71; hard target. |
>Domain of unknown function (DUF5026) (Length=82) LIVDSITKVFDTAQIKKGYLMYAKHRSWPDGRGGFVTAVTDKQITVQYHPGIANVTNHFFLPADEVAAGEWEVRWSADMI EV |
| 2814 |
PF13605 (DUF4141) |
 Estimated TM-score=0.71; hard target. |
>Domain of unknown function (DUF4141) (Length=53) RITMIICLCLLFAGRASAQWVVSDPGNLAQGIINASKNIIHTSKTATNMVSNF |
| 2815 |
PF18990 (DUF5723) |
 Estimated TM-score=0.71; easy target. |
>Family of unknown function (DUF5723) (Length=391) PAFRPERGYVSIPVIGSLGATYSSNGVAINDLFYPKGGKLVTFLDGSVETNAFLKRLKKNNQFNVDFNTSILSAGWYAGK GFWSADISVKGNASIRTPKTLFEFMKKGTGLEGETYDIRDINAYVDSYVEAAVGYSRPINERLTVGGKLKILLGVANMDA SIDHLRAEMYDNYWKITSTGKLSATMKGLSPTMDIDNKGEEYINDFDFDTPGVGGFGTAVDLGASYKLLENLTLSAALLD LGFINWSAGNTTAGAINGEEFNFNGFDLAIGENKDDIPSMSDQFEDLKDDINNLFHFKETGAKGRTTMLRATLNLGAEYS ILQNKLGFGLLSSTRFYRPKAYTEMTVSANYRPLDWLSASVSYSFIHSDFKTFGWALNFSPSWIHFFVGSD |
| 2816 |
PF12634 (Inp1) |
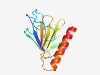 Estimated TM-score=0.71; hard target. |
>Inheritance of peroxisomes protein 1 (Length=130) ETLYSHPNVKIISFTATGRAYARSPGGPALSNVDPPSSLSWSSQLERTIAVGPFRIYRAPRSVAFLSCGSALQPILRKSQ CWCIDEVNSRFVLQIRRPNYWRIELPVDDPEDQQRAEELREVLDKILQFE |
| 2817 |
PF11256 (DUF3055) |
 Estimated TM-score=0.71; trivial target. |
>Protein of unknown function (DUF3055) (Length=80) LYDDTENTKTRFVSFLGENHRFDFAIVQTNRHYGKHLVLDMQGNRFAIIGHDDLEEEGYLEHAFQLNEHDAEELRSFLSE |
| 2818 |
PF06162 (PgaPase_1) |
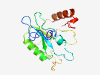 Estimated TM-score=0.71; easy target. |
>Putative pyroglutamyl peptidase PgaPase_1 (Length=164) MLSMCNKYPDFDDEERVRRVDDDFRDVITVFDAPFEGKSPSPAVIVFDELVDSDGSSKYLNFKMEQSYEKVDEIPEKIDA ERIRYAIHLGSHSQKNVLQIFQSAYSDGYTEEDKNGKVPVGGKVKCAETETGFRTKINCENVVTAVNEYMDSNREKFGEL RIET |
| 2819 |
PF14927 (Neurensin) |
 Estimated TM-score=0.71; hard target. |
>Neurensin (Length=130) RYGVRSYLHQFYEDCTASIWEYEDDFQIQRSPNRWNSVLWKVGLISGTVFLILGLTVLAVGFLVPPKIEAFGEGDFVMVD SQAIQFNGALDICKLSGAVLFCIGGMTMAVCLLMSAFAKSYSKEEKYLQQ |
| 2820 |
PF12545 (DUF3739) |
 Estimated TM-score=0.70; hard target. |
>Filamentous haemagglutinin family outer membrane protein (Length=111) GGDILAWSAEGDINAGRGSKTTQVYTPPKRVYDNYGQVTLSPSVPTTGAGIATLNPIPEVRPGNVDLIAPLGTIDAGEAG IRVSGNVNLAALQVVNAANIQVQGTSTGVPT |
| 2821 |
PF17255 (EbsA) |
 Estimated TM-score=0.70; hard target. |
>EbsA-like protein (Length=134) FGKIRYHWQPELSWAIIYWSIAIAPIFIGLSLLYERTEIPSQVFVLFAIFIVLVGIGFHRYFVIEEDGYLRIVSFNFLRR SKFPIEDIAKIEVTKSSVTIKFNNNHERIFYMRKWPKKYFLDALAIEPKFKGEV |
| 2822 |
PF16094 (PAC1) |
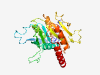 Estimated TM-score=0.70; easy target. |
>Proteasome assembly chaperone 4 (Length=287) MATFFGEVVAAPSRAGVDDEEEAAVEREETPEDREIRRELEKKREVHILWSSKLSASTGSFTDEQFPCSKFILAIGHNAV AFLSSFVLNSGCWEAVGSVKLWNEWCRTSNTTSVLPTDAFCLFYKLISDPTVLLCQCNCYVAEDQQFQWLEKVFGCMREE DLQVTILSTCPVTDYKTPESTLTLPSPFLKALKTKEFKEIVCCPLLEQPNIVQGLPAAVLSYCQVWQIPAVLYQCYTDVI KLDTVTIEAFKPVLSSKNLKSLVKDMSKSTEILKKLVTTNEIHNNIY |
| 2823 |
PF06396 (AGTRAP) |
 Estimated TM-score=0.70; hard target. |
>Angiotensin II, type I receptor-associated protein (AGTRAP) (Length=145) KMILLVHWLLTTWGCLVFLGPYAWANFTILALGVWAVAQRDSIDAISMFLIGLAVTIFLDIVHISIFYPRQNLTDTGRFG AGMAILSLLLKPFSCCLVYHMYRERGGELQLHAGFLGPSQDRSAYQTIDSPEAPSDPFSGPEGKN |
| 2824 |
PF06021 (Gly_acyl_tr_N) |
 Estimated TM-score=0.70; easy target. |
>Aralkyl acyl-CoA:amino acid N-acyltransferase (Length=205) MIHLQSAQKLQMLEKSLRMVLPESLKVYGTVFHMNQGNPFKLKALVDSWPDFNTVVIRPQEQEMTDDLDHYTNTYLIYSK DPEQSQEFLGSPAVINWKQHLQIQSSQSNLNRVIQNLVDINLSKVKHTQCNLYMVYDTAKKLAPSLVDVKNLSPKGDKPK PLDQEMFKLSSLDVTHAALVNQFWHFGGTERSQRFIERCIRTFPS |
| 2825 |
PF06939 (DUF1286) |
 Estimated TM-score=0.70; hard target. |
>Protein of unknown function (DUF1286) (Length=124) MKLRTHYVFSTGLLTFLDSVLFHEYFYYALILSGIVSVIGNSLIDRIGHKEIATRYGYIPVRTPLTHTIPRSVVWGIVSI IPVFILLLIYYGFSYHEYYFSLSNKVLMLILLNGVVVGPSHMLL |
| 2826 |
PF16998 (17kDa_Anti_2) |
 Estimated TM-score=0.70; hard target. |
>17 kDa outer membrane surface antigen (Length=114) GGMDILSSAKVDRSVATGTIPTAPVTTDSVSDETTVRNAVTSADLHKLNGQPVPWANASTGSAGVIDTIVENSASGQVCR QFRTTRHSYQGIANFSGRTCLLGQGEWQLLSFQQ |
| 2827 |
PF12987 (DUF3871) |
 Estimated TM-score=0.70; very hard target. |
>Domain of unknown function, B. Theta Gene description (DUF3871) (Length=318) FAEEATIIEETATPKRVNHFIEANTQEATLQHLKNDCITPVFAKDNELTVNHAAFIETVWEAANSYYNGETIGTPDIRVS HVIKGRIPEAIHKPANQLLESDKTIYYERAAFSIDIPTIYETVEGNKLHLSIVGVRAYNQMNLYSKKVPELFRLAIGFKN QVCCNMCIFTDGYKDDLRVSNTSELYRSALELFNNYNPAKHLHLMQQLGNTSMSEHQFAQILGKMRLYQCLPNGYQKRLP RMLLTDTQINSVAKAYINDENFGSFGNDLNMWKFYNLLTGANKSSYIDSFLDRSLNATEMAVGINAALHGDEDYKWFI |
| 2828 |
PF05212 (DUF707) |
 Estimated TM-score=0.70; easy target. |
>Protein of unknown function (DUF707) (Length=290) TNPRGAERLPPGIVNAESDLFLRRLWGLPSEDLTFKPKYLVTFTVGYEQKKNIDAAVKKFSKDFTILLFHYDGRTTEWDE FEWSKQAIHVSVHKQTKWWYAKRFLHPDIVAPYDYIFMWDEDLGVENFNAEEYLKLVRKHGLEISQPALEPSKSTKAVCW NMTRRREHSEVHKEAVEPGKCKYPLLPPCAAFVEIMAPVFSRNAWRCVWHMIQNEFVHGWGLDFAFRKCVEPAHEKIGVV DTQWIVHQGIPSLGNQGETQTTGKPAWRAVKERCGMEWRMFQGRLTNAEK |
| 2829 |
PF02040 (ArsB) |
 Estimated TM-score=0.70; trivial target. |
>Arsenical pump membrane protein (Length=423) LAVFIFLLTLVLVIWQPKNLSIGWSACGGAVLALIAGVVNFHDVLTVTGIVWNATLAFVAIILISLILDNIGFFEWAALH MAKAAKGYGVRMFVYVSLLGAIVAALFANDGAALILTPIVLAMVRALHFNEKLVFPFIIASGFIADTTSLPFVVSNLVNI VSADYFHITFIDYASRMVVPYLFSLLASIIVLYLFFRKSIPKRYDLTEVKKPVEAIKDQNMFRLSWYILGLLLIGYFASE FFSIPVSVVAGSIAIIFLIAAQKSPAVHTKKVVKEAPWAIVFFSIGMYVVVYGVRNAGLTDVLSDVIQAAADQGLFAGTI GMGFIAAILSSIMNNLPTVMIDALAIAGTDTHGMMREALIYANVIGSDLGPKITPIGSLATLLWLHVLSHKGMKISWGTY FKTGIILTIPTLLITLVGLYIWL |
| 2830 |
PF11872 (DUF3392) |
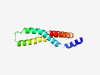 Estimated TM-score=0.70; hard target. |
>Protein of unknown function (DUF3392) (Length=104) LIEFLADSGQFIRPWLSEISTAMIACLLVVFGADINRLLRRQLSGTNFVIRTLVFILVNAFGYGFLIITVSPWLANQLSH IPSQWLICLVAATFIFIGTWAQRN |
| 2831 |
PF13032 (RNaseH_pPIWI_RE) |
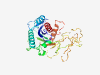 Estimated TM-score=0.70; easy target. |
>RNaseH domain of pPIWI_RE (Length=279) VAFHLVERRVDGPTGRAQFTPIAVLLRPDAPCVLGRTADTSEWMPYPDLLRILTGRVRGNDLRTSAQQSAVTAAFIRRTL ASLRGTPTLVLSHAQDVRKRWPWLTNGRLEVDRIGLDGGPAQRIGLYGKHLRVVRVADSGRDETPQWWAEREEREEELAG QQRGGFGMGLWVPDEPGAPGRVFYSTADKASTQTKLTNDDAKLTPHVNPAGRSAHRPTVNAWNPELLELTLACLQPGDDP EAWAAFVHQQRICKDDYRDVLGLPLALHLARLADEYALP |
| 2832 |
PF08350 (DUF1724) |
 Estimated TM-score=0.70; easy target. |
>Domain of unknown function (DUF1724) (Length=60) TILVCEDTIKPPSFSVTDTFLYMSLFDINGRYDHHDIMSFEDSALEWGERLFEYYQNLSS |
| 2833 |
PF12652 (CotJB) |
 Estimated TM-score=0.70; hard target. |
>CotJB protein (Length=77) ELLNHINQVSFAVDEVKLYLDTHPCDQEALVYFHEYSRKRNEALNEYARAYGPLTIDTASESCTERWNWINEPWPWQ |
| 2834 |
PF11660 (DUF3262) |
 Estimated TM-score=0.70; hard target. |
>Protein of unknown function (DUF3262) (Length=78) MNPSQQAAFEAAAGPGMSPSVLNLLCIGALLAVLFLWAAWGLVDVYRGWANDSVRAAKLGQFAVRAVILLVVCIWMFA |
| 2835 |
PF18656 (DUF5633) |
 Estimated TM-score=0.70; trivial target. |
>Family of unknown function (DUF5633) (Length=40) GFKTKAEAEDAAKKVLKEDKVNKSFTVTQGANSNYYFSLS |
| 2836 |
PF12788 (YmaF) |
 Estimated TM-score=0.70; very hard target. |
>YmaF family (Length=97) KNHNHEFESSTDYEEDDECELHNHRIAGVTGPPIRYGKTHIHKVAEFTDTFGDHFHEICDTTGPAIYLPGGKHIHLVKGR TTVADGHQHDYFFTTLI |
| 2837 |
PF10772 (DUF2597) |
 Estimated TM-score=0.70; easy target. |
>Protein of unknown function (DUF2597) (Length=134) MNFDITVGDMQVHVETVTLDITDNSAVSQTGGVPDGYVDGDVAASGEIEVDTTNFNLIVEAAKAAGSFRQLEPFDSLFYA KTPTDELRVEAFGCRLKISSLLNIDSKGGEKHKHKIPFDVTSPDFIRINGVPYL |
| 2838 |
PF14023 (DUF4239) |
 Estimated TM-score=0.70; easy target. |
>Protein of unknown function (DUF4239) (Length=220) FLPGVAIVLGTYFSLTLSILYNRFSQIQQTVTSEASLLALCCRNLLDMLCSNDIRKDCDKTAVQAAQCIADQVRTLVRDS RGRETMMVIYSDPYTRLLQIVAECRNEGIKLDEALVANVRSNVADLCKLRAHRISYEGLALAPTHFDVMTFLCGLLLTGY ALGTLATACPNDGTPAGLAQVLFSALVVCYVLFYEMCYDLNRPFDGIYQLRRSGAAMHLL |
| 2839 |
PF09573 (RE_TaqI) |
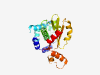 Estimated TM-score=0.70; very hard target. |
>TaqI restriction endonuclease (Length=224) LEQFETFLSTVHLATYRTRYLPIKIVEMDLPKDIHAIEKLYEVYWDKKKFLDFDDFYKEYLDSKNSEIDSFRQKITMCSD CFYRGLPARIYRTWASIITQIHAGYVAESVFGEGSVKMSAELDHKGADFQVHYHSRTLNYQVKKKSLGREVRQEKPKAKN QLAGDFVDIKYEVPSSDYFENPKRKDGEYKLPYKRFIENKELKRFPNGFVVFTTYAFERKKLEI |
| 2840 |
PF04235 (DUF418) |
 Estimated TM-score=0.70; hard target. |
>Protein of unknown function (DUF418) (Length=164) ARKKVFEDLEGNRPFFRRLLVWGLIIGVTGNIAFASLIGPDSPRYIPSLKVLVATLGQSFGAPALSLGYVAALVLLSCRS AWQQRLAPLAQVGRMALTNYLGQSIICTLIFYGYGLGLFGQVDKAAGLLLAVVIYAAQVWFSNWWLKRFQYGPMEWLWRS LTYL |
| 2841 |
PF10849 (DUF2654) |
 Estimated TM-score=0.70; hard target. |
>Protein of unknown function (DUF2654) (Length=70) AERKANKLLSKNKRELNRLYKHAQIAAENNNFAQYEYAIKKSRDILKQPYNDELISILWKTTRSQIEDMI |
| 2842 |
PF04526 (DUF568) |
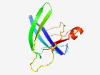 Estimated TM-score=0.70; easy target. |
>Protein of unknown function (DUF568) (Length=100) MAGSQALVAFKDSNGSMTVRTYNISSYSSIVPSNLSFAVTEMAADSSGGMMRIFAKVTLPKKGATTVNQVWQVGPSLTNG VLDKHEFQPANLNSKGTLDL |
| 2843 |
PF15584 (Imm72) |
 Estimated TM-score=0.70; very hard target. |
>Immunity protein 72 (Length=81) PKNESSTGEVNSGEEIPVEGIWEPWFPGDEVGCPNYFLKGSIAHQYLLEGTNDEYDVRWRLLWEDTRYQGGSIPEEESTY F |
| 2844 |
PF14457 (Prok-E2_A) |
 Estimated TM-score=0.70; hard target. |
>Prokaryotic E2 family A (Length=165) ERFLAAALRHPECRGGRLISVDAGGSRIELDLNVEMPLAFKVDGASPNGVRVVETVNVRLWPSYPWSSPSFYLRMDFPRD LPHVQPGPVTEPPRPCLIDGNQREYFFQFGLVELGIFNLVHQLVLWLQRAAEGTLIHHGRGWEPTLRCDLNDVIALNAEA CRAVV |
| 2845 |
PF16371 (MetallophosN) |
 Estimated TM-score=0.70; easy target. |
>N terminal of Calcineurin-like phosphoesterase (Length=77) GYVHDSEKPLSGVLVTNGYSFATTDENGAYYLPSDADAEFVYLVTPAGYNADCRDGLTRFYQTIDKNGGIYRADFQL |
| 2846 |
PF06691 (DUF1189) |
 Estimated TM-score=0.70; hard target. |
>Protein of unknown function (DUF1189) (Length=244) FVQLWKSLYSPKDIARFRFQGIGKTIGYVFLLTFLSILPMAYYLTTSFVEGIRNTSALLQNELPSFTIENGQLHSSAKEP IHIDQHGFTIIFDSTGQVTKEDMERFNNAIGLLKHEVVLIANHQTQYYSYSTFPDVKITDEDVHSFIETMQSLLPVFIPL LFIIIYLFASASKFIGICILAFFGLILRSTLDKKLQYRHTWIMSAYAVTLSTVFFAVMEALQTVVPYAPFIHWFVSIAVL FLAM |
| 2847 |
PF12260 (PIP49_C) |
 Estimated TM-score=0.70; easy target. |
>Protein-kinase domain of FAM69 (Length=202) AWALLQLDEFLLMVILQDKEHTPKLLGFCGDLYVTERVEYTSLYGMSLPWIIELFIPSGFRRSMDQWFTPSWPRKAKIAI GLLEFVEDIFHGPYGNFLMCDTSAKNLGYNDKYDLKMMDMRKIVPEMNLKEIIKDRQCESDLDCIYGTDCRTLCDQSKMR CTTEVIQPNLAKACQLLKDYLLRGAPSDIHEELEKQLYLCIA |
| 2848 |
PF06213 (CobT) |
 Estimated TM-score=0.70; very hard target. |
>Cobalamin biosynthesis protein CobT (Length=272) EAPAEPFKRSVAGCLRAIAKTPELEVTFASDRPALTGERARLAEPPRKMSAADAAIVRGQADSMALRLACHDAVVHRRLA PEGQQARAVFDAVEQSRVESIGSNRMAGVASNITAMLEDRYHRGAYHEVTDRADAPLEDAVAMIVRERLTGLKPPPAAQK IVDLWRPWVEEKAGKDLDKLASNLEDQRHFARSVREMLASLDMAEDSSLDPEDSEEDEESNEQSQDKEQQEGESEEQSAA ERAEMERSEEATEEMEEGQSETSEGPTGEMPD |
| 2849 |
PF10622 (Ehbp) |
 Estimated TM-score=0.70; hard target. |
>Energy-converting hydrogenase B subunit P (EhbP) (Length=77) MQPKHMVSLGGYIVETQFPYRNLIVINKTSEPIKIEIPVFDESWIEEHRELGLEVIPVSKEDNYLKMWKRAHAELDK |
| 2850 |
PF03649 (UPF0014) |
 Estimated TM-score=0.70; hard target. |
>Uncharacterised protein family (UPF0014) (Length=220) VAATAVVAMAVALSFAQRLGLEREMLYSVARAFLQLSVVGFVLHFIFTQKNAPWILLAYLFMVTVAGYTAGQRAKHVPRG KYIACVSILVGTAITMLVLVVLKVFPFTPRYIIPLAGMLVGCAMTNTGVTMKKLREDVKVQRNMVETALALGATPREATL QQVRRSLGIALAPVIDNAKTVGLIALPGAMTGLIMGGASPLEAIQLQIVVKNMVMAANTP |
| 2851 |
PF07810 (TMC) |
 Estimated TM-score=0.70; easy target. |
>TMC domain (Length=111) CWETYVGQQLYKLILTDFAVQFVTTFFINLPRAFLARHTNSRCFKIIGEQDFYLPKHVLDIVYTQTIIWMGSFFCPFLPI IGTIFYFLIFYIKKFTCLTNCTPSPVVYKAS |
| 2852 |
PF08743 (Nse4_C) |
 Estimated TM-score=0.70; hard target. |
>Nse4 C-terminal (Length=91) IDLLRFVINPRSFGQTVENMFYVSFLIREGSVKLEFDEDGLPAIAPVRKNSSAEPSRPKATTRHQAIMSIDMATWRDIID AFDIKEPMIPH |
| 2853 |
PF15646 (Tox-REase-2) |
 Estimated TM-score=0.70; very hard target. |
>Restriction endonuclease fold toxin 2 (Length=134) PDNAYQLRIAGYPEREVPLAGQRRGLMVDGIRPADGHLVEAKHVRDPDCTKKSFRSLDRVNETLVKPVKINSQGKIAWDP MVDSMYRGDQRELNRYKQAMDNPANNEIRGLEIVTNGKDQAAYWQSMMAMTGTP |
| 2854 |
PF17240 (DUF5313) |
 Estimated TM-score=0.70; hard target. |
>Family of unknown function (DUF5313) (Length=123) DRPNLLQYIAYSYGRRLPDSMREWVAHDLADHGAVRRHMIRMAIPPALVLAPFWLLPASLYVHIEMTVPIYIWSLLMALA LNKVWRRHRLAQHGLDPNLVDEIRYKKQAHIHEDYIRRFGPRP |
| 2855 |
PF07129 (DUF1381) |
 Estimated TM-score=0.70; hard target. |
>Protein of unknown function (DUF1381) (Length=44) TQYLVTTFKDSSGLPHEHFTAARDNQTFTVVEAESKEEAKEKYE |
| 2856 |
PF07441 (BofA) |
 Estimated TM-score=0.70; hard target. |
>SigmaK-factor processing regulatory protein BofA (Length=75) IAIVGLYVIVKVFSWPIKILFKLIINAVLGVVLLVIVNLIGSYFGFSIGINAVTALIAGFLGIPGVIFLIIFKLF |
| 2857 |
PF06580 (His_kinase) |
 Estimated TM-score=0.70; easy target. |
>Histidine kinase (Length=80) ARLDALKLQLNPHFFFNSLASVRSLISEAPRRAKKMVARLARLLRKTLQASDEETVSLREELSTTQTYLQLEKVRFEDRL |
| 2858 |
PF12482 (DUF3701) |
 Estimated TM-score=0.70; easy target. |
>Phage integrase protein (Length=92) TLRNLPAPAPLITDAVEVWLPPRIAEALHAHGIRTLADLTVRIPRRRRWWSAIAGLGVAGARRIEAFFAAHPALTERARA LIVAVPSGSIVP |
| 2859 |
PF03077 (VacA2) |
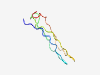 Estimated TM-score=0.70; hard target. |
>Putative vacuolating cytotoxin (Length=60) GYIGFITGVFKARDIFITGAVGSGNEWKTGGGAILVFESSNELTTNGAYFQNNRAGTQNS |
| 2860 |
PF04342 (DMT_6) |
 Estimated TM-score=0.70; hard target. |
>Putative member of DMT superfamily (DUF486) (Length=104) TILLLTISNIFMTFAWYGHLKYKDSPLWIVIVASWGIAFFEYCFQVPANRIGHYEFNAAQLKTIQEVITLVVFCVFSVLY LKEPLKWNYLAGFGLMIGAVFLIF |
| 2861 |
PF10112 (Halogen_Hydrol) |
 Estimated TM-score=0.70; hard target. |
>5-bromo-4-chloroindolyl phosphate hydrolysis protein (Length=189) RIIIAIPVAVLVWVISYFGFEQAFFPSSIFAILGGAIIYLIIKWISTRKFLKRHQLTRKELTYIVQNLKEAKKKIHRLQK VFFNVRNIGAFKQMLDLIRLVKRIYSIIKKEPRRFYQAEKFFFYHLDSIVELSEKYAFLAAQPVKNNELYFSLKDTRNTL KELKRTIENDLYEVLSKDIDHLEFELNVA |
| 2862 |
PF16397 (DUF5006) |
 Estimated TM-score=0.70; easy target. |
>Domain of unknown function (DUF5006) (Length=263) KNILYACLPLGIWAMLLTGCKDDDQIAPLPKPVPLTLALESNTLVMGETLNMTFSVKDEQGAGLAANEDFDIYLAVVEGT TDVSKAVFNNFPEMVTFPKGETDLEISVPVKSSGITKSVLATLTAFARGYKMDGSEQVVKVSDYYRTTISIKGNSDLVVR EGDTFILQMKVEVPAKEDIVVTVTPGAGESDFYENLPSTLTIAAGELSVESEPITMLADGYPFGDRKLTFKFTSESVHHP LLSEEMNLTQQDIDTPLGSELED |
| 2863 |
PF15524 (Ntox17) |
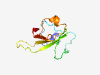 Estimated TM-score=0.70; very hard target. |
>Novel toxin 17 (Length=94) WSITARIQYAKLPRQGRIRYIPPKNYSPSAPLPKGPNNGYLDKFGNEWTKGPSRTKGQEFEWDVQLSKTGREQLGWASRD GKHLNISIDGKITH |
| 2864 |
PF15635 (Tox-GHH2) |
 Estimated TM-score=0.70; very hard target. |
>GHH signature containing HNH/Endo VII superfamily nuclease toxin 2 (Length=118) QTAHHLIEASALHDKGRGGKGSVPLKGISNYSENKAPCVCAEGVNQNVGTHGLMHTFQSAAAAKSRSGILQLSNGSSISA KKTTYGTAKRQSMAAMGKVFPQSKCSKECLSAQLDNYH |
| 2865 |
PF07812 (TfuA) |
 Estimated TM-score=0.70; hard target. |
>TfuA-like protein (Length=120) DGYFETVPTVWHKEILWAMSQGIHVYGAASIGALRAAELADFGMKGVGHIYRQYRTGELTDDDEVAVLHGPAEVDYVQVT EAMVNVRATLEHALQSGIVEPDVASALVEVAKSLFYKDRT |
| 2866 |
PF13069 (DUF3933) |
 Estimated TM-score=0.70; hard target. |
>Protein of unknown function (DUF3933) (Length=53) MKQYVICQVIDGMKYLAAYAETRQEAIEKAELLGLRTGGRYVVVTAEEAEGLQ |
| 2867 |
PF11859 (DUF3379) |
 Estimated TM-score=0.70; hard target. |
>Protein of unknown function (DUF3379) (Length=233) MDELKFRRQAYEDPHNQDPEFVAQMQESAENQAFVAELKSLDAKLTHALKVDVPEDLADKLILRQQLQQHHKQRRQTGFL VAMAASIAFIVGISFSLLRLGPVDLAEHALAHVYHEGVALQVDQNVNFSQVNAQLASLKNLGHAKFTEQPGKVYYTSYCD FQGVKSLHLVLQAEKGKVTLFIVPIEKRMVLDNTFADGKYQGMGFEAGDAYILLVGEDKTDLSFVKDEIKNTF |
| 2868 |
PF09774 (Cid2) |
 Estimated TM-score=0.70; very hard target. |
>Caffeine-induced death protein 2 (Length=157) CFNERVLRDFLRLSRSAIDDSITQNLNALITPARVGFDPSSTAVRQTGKKTIDPAACQSFKDNVLFPSWQTRSDVLNYCA GVATSPDPEDPDLILRETESAKDRERVVDERLDPYSARFFPKEARTESLAYLIRNERSVEQIIRARTWGLVTERCGD |
| 2869 |
PF06034 (DUF919) |
 Estimated TM-score=0.70; easy target. |
>Nucleopolyhedrovirus protein of unknown function (DUF919) (Length=62) SKDEDLRRQLNQILQAKRQLTIQMEHWERIKRITKDPKEVQDIESKLLKMRMDFLNFSTDKF |
| 2870 |
PF16506 (DiSB-ORF2_chro) |
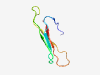 Estimated TM-score=0.70; hard target. |
>Putative virion glycoprotein of insect viruses (Length=51) SPYMFDRSCLNVYRTNDYLFGECTLPPNCSEPSVVKLDKTFYGQETVVCHS |
| 2871 |
PF16947 (Ferredoxin_N) |
 Estimated TM-score=0.70; hard target. |
>N-terminal region of 4Fe-4S ferredoxin iron-sulfur binding (Length=65) HPLGESWQDDLEEMLDDTEYDTELGMEMGRDAMRVTKGELSEAEFHERYHDDVVAEFGEDDRPIA |
| 2872 |
PF14351 (DUF4401) |
 Estimated TM-score=0.70; hard target. |
>Domain of unknown function (DUF4401) (Length=314) PWYMRIFAGIGAWFAAFMFLGFLAITNIITNEFGALVVGLGLCAVAIGINRTNQASTFLRQLALAISITGQALVLFGWTI IVESVIVAALGMIVLEIALFVAHRDFSQRLISTLGVITAIHVIAGDLNNWVESFNYMYLLLSVMIISFSLLMWFEAKIHE DQRWSSASPIAYGMLLFMAISSLLINFNWAEFFSRGSFFILPALVGIICLGTTIWMIINEFEYKLGFIELGCIAFGLLFI AFAGINTPAIIIALLIILLSYWRGYPVMFGFGCLALIGALSLYYYNLNISLLYKSIILFGTGMMLLAIRALVLK |
| 2873 |
PF13031 (DUF3892) |
 Estimated TM-score=0.70; hard target. |
>Protein of unknown function (DUF3892) (Length=69) KIVKVKKDHSGDITDVMMENGNVYSINEAIMMAKDNLIEGVNVGVSKSGREYIRSNPNGTEKDNLDNLP |
| 2874 |
PF14617 (CMS1) |
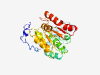 Estimated TM-score=0.70; easy target. |
>U3-containing 90S pre-ribosomal complex subunit (Length=268) STTDRRKRKREREKLNKQKGKKSKSEGDEIAPQLLEHRSKKETRKGGEVKSTATDVPGGETEPVNEDMARMDPKLLADYV SQRLKRFEKDLSSVELEDRYISESAFLDTTAFTPPRTLRNLPAYLESFSKFALIKSAATPGTPHTLILTLAAVRATDLAR AVRKYQTKDSLVAKLFAKHIKLKDSITTCQTSRIGIGVGTPGRILDLLKEGVLKVDELKNVVIDASYVDKKGFGAFDIRE TQKGIMEILGFAGVKARFERGEGRVMFY |
| 2875 |
PF09333 (ATG_C) |
 Estimated TM-score=0.70; hard target. |
>Autophagy-related protein C terminal domain (Length=96) VSKYADQPNDVREGVEAAYKSLGKNLTSAAQTILAVPMEVYERSGNEGAARKVIRAVPIAVLKPMIGATEAVSKTLLGLR NTLDPENLAETEEKYK |
| 2876 |
PF12474 (PKK) |
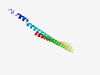 Estimated TM-score=0.70; hard target. |
>Polo kinase kinase (Length=137) QLAKEQQERRFEQEHIALQRTYEADMDGLTRQHRQLIEKTEQQQENDLRATSKKIRAEQERDLKLFRDGLKQEIKLLKQE IDLLPKDRRKVEFPKRKAAMEAEHEEKERAFLSSLSESHEIALRRLSEKHRDRLATI |
| 2877 |
PF13991 (BssS) |
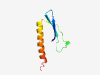 Estimated TM-score=0.70; hard target. |
>BssS protein family (Length=71) HPLVGWDISTVDSYDAMMIRLHYLSSKDQTPENAQVDRTLWLTTDVARQLIHILEAGIEKIESNEYEYSDH |
| 2878 |
PF06045 (Rhamnogal_lyase) |
 Estimated TM-score=0.70; easy target. |
>Rhamnogalacturonate lyase family (Length=219) ANVNKSSTYKLRVALASATFSELQVIMDNGIVRVNLSNPEGIVTGIQYNGLDNLLEVLNEESNRGYWDIVWDQGGEKRTE KGKKGKGTLDRLEATNFTVIVKNEEQVELSFTRTWNSSLEGKFAPLNIDKRFIMLAGSSGFYTYAIYEHLKEWPAFDLDN TRIAFKLRKDKFCYMAVADNRQRFMPLPDDRLPPRGEVLAYPEAVRLVDPLEPEFKGEV |
| 2879 |
PF16424 (DUF5021) |
 Estimated TM-score=0.70; very hard target. |
>Domain of unknown function (DUF5021) (Length=178) VTSANSTAASIKKQIDNFLTDADTAGYGMKQSSAAKANITFKINADGEWEASVVTGTYTGGAAGGALTDAFKTGGSIQWG ASAADITKDTPKSSAGDATALLTIDLASVFPDVKSSYIYAYCEGGKTLYVAYTADGNTKPTSMPGEADFKKGTYTWDGNT AGITSDGITLGTAPALSL |
| 2880 |
PF17184 (Rit1_C) |
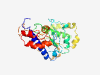 Estimated TM-score=0.70; very hard target. |
>Rit1 N-terminal domain (Length=258) LKRSNLSITNRLHSIQEDAYFVSEVTSAFNDRPLIANERCGSWYIAPQNKRGSAYFKSTDGHTGQWKFSTRRLNVHLLSI IGENDGCIIVDSTRRGKRMPDALSKTVPVWCCVLNRALFPNATERHRLYTPPNVVSESEHAQVEARIEGFVESLKRLDID LEGLRGQVKRPLRPMWVTQDSQLVPTNEVFEDFHPVICCTCSRRVVGGEMSEGGYIQGAGDDTENWALGLTPPVFWEHEK ELLSTPEADLRDVVLALV |
| 2881 |
PF07394 (DUF1501) |
 Estimated TM-score=0.70; easy target. |
>Protein of unknown function (DUF1501) (Length=423) AKRVIFLFMAGAPSQIDLFDYKPTLQRYDGQPCPESLLQGERFAFIKGRPTLLGSPYKFSKHGQSGQEISELLPHIAQIA DDICIIRSMQTDQFNHAPAQIYMNTGHQLPGRPSMGSWLTYGLGTENRDLPGFVVLISGANRPDGGHACWSSGFLPTVYQ GVELRSKGDPVLYVSNPEGIDSETRRETVEAISDLNRMHLEAVGDPEIETRINAYELAYRMQTSVPELMDISKEPEHIHQ LYGTTPGKPSFANNCLLARRLVERGVRFVQLYHRGWDHHGTGEGDSISKALPRLCSEVDRASAALIIDLKQRGLLEDTLV IWGGEFGRTPMKEGRGGSTYLGRDHHPRAFTIWMAGGGIKPGITYGATDELGYQVVENPVHVHDLHATILHLLGIDHTRL TYWYQGRNFRLTDVAGRVIEGII |
| 2882 |
PF11067 (DUF2868) |
 Estimated TM-score=0.70; very hard target. |
>Protein of unknown function (DUF2868) (Length=297) ARDAQAAQLAPALLLMLQRQRLSRWGLGVLVQGFWLLTLITALLTLLALLATRRYGFVWESTLLGGDSFIAMTQAIGSLP ALLGFSLPDAELIRASGDSALANESARLSWAGWLVGVLVVYGILPRLALLLLCLWRWRRGCAALSLDLSLPGYSLLRERL QPSSERLGVCDAAPAQLHQPQAGNHSEGSQGALLVAIELDPQRSWPPVLPKGIGDAGVIDSREQRNQLLDQLNRFPPARL LIACDPRRSPDRGSLALLGELARSAAATRIWLLPPPPGEALDSARLGDWHRALEQLQ |
| 2883 |
PF04246 (RseC_MucC) |
 Estimated TM-score=0.70; hard target. |
>Positive regulator of sigma(E), RseC/MucC (Length=135) VVAVEGAFAWVETRRRTACGSCSAQAGCGTSVLDKMFGQRMNRIRALNQAEARVGDEVIIGLSESALVRGSFAVYAIPLV GLFVGAGLATAMSGAEGEGLAILGGVLGLAAGFLAVRRFSRRIQTDRNYQPVVLR |
| 2884 |
PF12158 (DUF3592) |
 Estimated TM-score=0.70; easy target. |
>Protein of unknown function (DUF3592) (Length=135) LILAVLLIGTTVAGAVLYELDVREAGNLRSKWPTTQGTILSSTILRDFENSETAFIIIRFAYEAKGTRYSAQQAWREWLT RTHQYSEGSSVTVYYNPAKPSEAVIEPREQSPGSSLIGFGLFAMALVAIIAGIWI |
| 2885 |
PF14772 (NYD-SP28) |
 Estimated TM-score=0.70; hard target. |
>Sperm tail (Length=101) ATDIRETHRRVEEDEVKRQRLEKLENEAKTSQCKFEEITSKWGEGKKKKIPQDLWEMLNTQQQQCAQLIEEKNKLINELQ QELKGKDDQYVKDLKKQGDDI |
| 2886 |
PF14012 (DUF4229) |
 Estimated TM-score=0.70; hard target. |
>Protein of unknown function (DUF4229) (Length=68) VLYGLARLLLFIVLTAVIQGLSVLVGITFPLMISALLALIIAFPLSMFLFKGLRQRVTEELAEWDRQR |
| 2887 |
PF03217 (SlpA) |
 Estimated TM-score=0.70; easy target. |
>Surface layer protein A domain (Length=52) KIMSKSYVYTSAGKRTGAYKSAYTYVYVIGGIVKIGSNNYYKIGTNQYIKVN |
| 2888 |
PF04917 (Shufflon_N) |
 Estimated TM-score=0.70; very hard target. |
>Bacterial shufflon protein, N-terminal constant region (Length=329) DYIQTKGWQTEARLVSNWTSAARSYIGKNYTTLQGSSTTTTPAVITTTMLKNTGFLPSGFTETNSEGQRLQAYVVRNAQN PELLQAMVVSSGGTPYPVKALIQMAKDITTGLGGYIQDGKTATGALRSWSVALSNYGAKSGNGHIAVLLSTDELSGAAED TDRLYRFQVNGRPDLNKMHTAIDMGSNNLNNVGAVNAQTGNFSGNVNGVNGTFSGQVKGNSGNFDVNVTAGGDIRSNNGW LITRNSKGWLNETHGGGFYMSDGSWVRSVNNKGIYTGGQVKGGTVRADGRLYTGEYLQLERTAVAGASCSPNGLVGRDNT GAILSCQSG |
| 2889 |
PF08320 (PIG-X) |
 Estimated TM-score=0.70; very hard target. |
>PIG-X / PBN1 (Length=212) PTGLHPKLDITFPIEDLKSPSPEKTCSLHAYMTTPSTLFLDRYQFEDKLFLASQNLVALRVLSGEEDLEAPNWVVKQWGS IALVELASPTSTEHPKGNWTVTIPMHLRYLNATASETGHTTIDAPWPVVFWACEAEEGLKMSTNPFDRINLGYDGLFGPK TMFYHVPANPENGRLVEQLQVPVLDPTQATWVPLGTLLAVVLGFAWICWKLF |
| 2890 |
PF10443 (RNA12) |
 Estimated TM-score=0.70; easy target. |
>RNA12 protein (Length=432) RRDLIEQIQNWLMESSDTFIVVQGPRGSGKKELVLQQTLEGRKNVLVIDCKPIVEARGEGGTIKNMATAVGYRPVFSWAN SMSSLVDLAVQSTTGVKSGFSETLETQVVKILHTAASALKDVSLASRHKNDKDADLSDDAYLDAHPEKRPVVVIDNFLHK NEESSIIYEKMAQWAAMLVQNNVAHVIFLTNDTSYSKSLSRVMPDRVYRQVALGDLSPDVAKKFVASRLEVDDTEKQKDS DSEKQQETPRSDLSELDDCIDLLGGRLTDLEFLARRIRGGQSPKHAVDDIINQSATEIVKMYLLGGKVDAEERKWSTEQA WYLIKSLVAQQSLRFNEVLLSDTFASSQTSRAASGELALESLTNSELITVKTHYGRPQTITAGKPMYRAAFKVLAEDPVL RAKMDLAVLTEMAKVEAKTIDKVENELALLGA |
| 2891 |
PF13845 (Septum_form) |
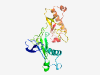 Estimated TM-score=0.70; very hard target. |
>Septum formation (Length=241) AGQCLDWKTSSDGTVSDFQTVDCAKSHRFEVSHRENLATYPASEFGSNAGMPSRKRQAELSNELCKQPTLSYLGGKFDPT GRFTIAPILPPQKSWADGDRTLLCGLQVTGNDGHASTITGKAADQDQSPVFKPGECVAVTEKKDDNQNKQTDGDTSVDAT AQVVDCNADHQYEVTDIVDLGEKFKEYPSTEDQNEYLNKRCTDSVTAFIGGDDKLYNSTLEPFWTTLTEDSWKGGSRNVN C |
| 2892 |
PF06819 (Arc_PepC) |
 Estimated TM-score=0.70; very hard target. |
>Archaeal Peptidase A24 C-terminal Domain (Length=111) LMISLTVIEIIKKLLTTVSREALQDDYNISELREGMIPAYNLYEKYDKVYTDDKSFLSKFKEAMKTGDVTGLTAPKGKLL ISTMAAGLTDKDVELLEDLLNKGKINDSFRV |
| 2893 |
PF08268 (FBA_3) |
 Estimated TM-score=0.70; easy target. |
>F-box associated domain (Length=124) ICINGILYYLALDSDGRYFMVVCFDVRSEKFKLVDIECRFDQLVNYKGKLCGIDLKNAYYGGFPLELRMWVLEDVQKREW SKYVYTLRDDNKVVKVNYNLSVSGMTATGDIVLLLNNASNPYYV |
| 2894 |
PF13224 (DUF4032) |
 Estimated TM-score=0.70; very hard target. |
>Domain of unknown function (DUF4032) (Length=163) ELWSALTDEESFGADERWRVAARIDRLNHLGFDVGELEITTDIDGTSVQIQPKVVDAGHHSRRLMRLTGLDVQENQARRL LNDLDSYRAAMDRQNDDEEFVAHDWLSGVFEPTVRAVPRDQRRKLEPAQLFHEVLDHRWFISEQQKRDVPMPEAVESYVE NVL |
| 2895 |
PF18923 (DUF5673) |
 Estimated TM-score=0.70; easy target. |
>Domain of unknown function (DUF5673) (Length=63) NKSGLTEKGILLNSYVAIWQKIESYSLDEEASRYVLLYKTNAGVRRIYFKKENIEEVRKYLSR |
| 2896 |
PF11190 (DUF2976) |
 Estimated TM-score=0.70; hard target. |
>Protein of unknown function (DUF2976) (Length=87) LPTMEPPSSGGGGGLFGQIKGYSQDGVVLGGLIISAIAFIVVANAAISTFSEIQDGKKTWTQFGAIVVVGVILLVAVIWL LTKSASI |
| 2897 |
PF09855 (zinc_ribbon_13) |
 Estimated TM-score=0.70; hard target. |
>Nucleic-acid-binding protein containing Zn-ribbon domain (DUF2082) (Length=62) CPKCGNTEYESDQFQATGGNFSKIFDVQNKKFITISCKKCGYTELYKAQSSAGWNILDFLIG |
| 2898 |
PF10601 (zf-LITAF-like) |
 Estimated TM-score=0.70; hard target. |
>LITAF-like zinc ribbon domain (Length=69) EPVTTTCPHCGSNITTTVNQINGFAPWIVCGGMALIGCMLGCCLIPLCINTLKDTEHCCPSCQQVIGRH |
| 2899 |
PF07762 (DUF1618) |
 Estimated TM-score=0.70; very hard target. |
>Protein of unknown function (DUF1618) (Length=136) WVDLWRNIIFCDVLAERPKLSYLNLPPALRPRPNMGAGDARSHRNIAVLGNTIKYVEMLPHPDRSSSKPASHRWEVAAWS IQKANRSWPKDWHTECNLDSTHIKVDSGTTAAALPTLSTLHVGLPTVSLQNDAIIY |
| 2900 |
PF03773 (ArsP_1) |
 Estimated TM-score=0.70; hard target. |
>Predicted permease (Length=299) HLGSAVNFFIYDTIKIFILLATLIFVISFIRTYIPPNKVKETLEKRHRYTGNFIAALVGIITPFCSCSAVPLFIGFVEAG VPLGATFSFLISSPMINEIAIILLLGLFGWQITAFYIVSGFIIAVLGGILIGKLKMETELEDYVYETLEKMRALGVADVE LPKPTLRERYVIAKNEMKDILRRVSPYIVIAIAIGGWIHGYLPEDFLLQYAGADNIFAVPMAVIIGVPLYSNAAGTIPLI SALIEKGMAAGTALALMMSITALSLPEMIILRKVMKPKLLATFIAILAVSITLTGYIFN |
[an error occurred while processing this directive]
zhanglab![]() zhanggroup.org
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218