

Go to page (83 pages in total): [First page] << < 23 24 25 26 27 28 29 30 31 32 33 > >> [Last page] [All entries in one page].
| # | Pfam ID | D-I-TASSER models ranked by estimated TM-score (click to view) |
Full name and representative sequence |
| 2701 |
PF09543 (DUF2379) |
 Estimated TM-score=0.71; hard target. |
>Protein of unknown function (DUF2379) (Length=118) ELDWEPIRALGQRVIERGEPLELSDEVRALLQRSAEEVALSPEDTANALRSVPTATTLLEEITRRIREGSDRFWNAELRA SDLQDAGDLDGAGRLMEEVLAVEVVPHYRRIAESQLRK |
| 2702 |
PF10307 (HAD_SAK_1) |
 Estimated TM-score=0.71; hard target. |
>HAD domain family 1 in Swiss Army Knife RNA repair proteins (Length=204) TPLPNPQLWNGPTIGTLSNQETFVNGGWWHDSRILAATGQGIDVEEKRAWEGWWNETIVDLVRLSMQQKDALCVLLTGRS EKGFSELIQKMVKAKGLEFDMIGLKPAVSPTNQKFNSTMHFKQLFLRGLMETYKQADEIRVYEDRPKHTKGFRDFFAEFN REQSSAPTRGPITAEVIQVADCSTTLDPVAEVAEIQHMINIHNE |
| 2703 |
PF07587 (PSD1) |
 Estimated TM-score=0.71; very hard target. |
>Protein of unknown function (DUF1553) (Length=260) NRLGLAQWLVHPEHPLTARVTVNRMWQHHFGTGLVKTAEDFGVQGEYPSHPALLDWLAVDFIESGWDVKRLHKLLVMSAT YQQSSRVDAAKLAADPENRLLSRGARFRLDAEVIRDQALAVSGLLVPEIGGKSVRPYQPPGLWKPVGFGGSNTSVFKQDT GDKLYRRSMYTFWKRTSPPPSMTAFDAPDRETCQVRRARTNTPLQALVLMNDVQFVEAARKFAECVVRQGGSSVEERVTF AYRSLLSRRPTASELASVTK |
| 2704 |
PF09852 (DUF2079) |
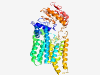 Estimated TM-score=0.71; easy target. |
>Predicted membrane protein (DUF2079) (Length=485) RYYSFYASYDQGIFNQVFWNSMHGHFFQSSLSSALSTNVVHQGQVSEVYYHRLGQHFTPALLLWLPIYVLFPYPITLTVL QVTLISVSGLVLYILARQHLQPQLSAMMSISFYGANAVIGPTLANFHDICQIPLFVFSLLLAMEKRWWSLFWILAVLTLA VREDSGVVLFGVGFYLILSKRYPKIGLAICTLSFSYMIALTNLIMPVFSEDISQRFMMERFGQYTEGDQASTLEIIWGMM SNPWRLIKEIFTPFFGTLKYLLGQWLPLAFVPAVAPAAWMIAGFPLLKLFSAKGLSVLSITIRYAMTVVPGLFYGAILWW GKRESEVKNSPVPSPKFLRFWVVCICLSLFFTFTSNPNRTFYFLVPDSVKPWVYIPANEQWQHVSQMRPLLDKIPDDASV AATTYIVPHLSSRRAILRFPRMQFRNDAKKIEKVQYIIVDLWRLNRYRVAFKSDRQRLETIVPRIEELYNSGEYGITDFG DGVVL |
| 2705 |
PF05604 (DUF776) |
 Estimated TM-score=0.71; very hard target. |
>Protein of unknown function (DUF776) (Length=180) MKSEAKDGEEESLQTAFKKLRVDASGSIASLSVGEGTSVRASVRAAVDDTKPKTTCPSKDSWHGSTRKSSRGAVRTQRRR RSKSPVLHPPKFIHCSTIASPSSSQLKHKSQTDTPDGSNGLGVSTPKEFSVGECSTSLDANHTGAVVEPLGTSVPRLPLE IKTEDSSDATQVSQATLKAN |
| 2706 |
PF15460 (SAS4) |
 Estimated TM-score=0.71; hard target. |
>Something about silencing, SAS, complex subunit 4 (Length=95) ALPDALYLPVHKRAERLERSIRNTEKGRAQHEKDQIVRLLEGLQGHDWLRIMGVSGVTESKKKSFEPARRHFIKGCQAII DKFRQWAQEEKRRKA |
| 2707 |
PF07841 (DM4_12) |
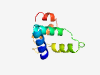 Estimated TM-score=0.71; hard target. |
>DM4/DM12 family (Length=83) RTYFYDQLEERMELYGFNASGCMERLICEVSELPLHEHNGVFGDVLSVIFRPSSSVREALPVSYYEAESRGSIEGCQQYR AFC |
| 2708 |
PF02411 (MerT) |
 Estimated TM-score=0.71; hard target. |
>MerT mercuric transport protein (Length=116) MSEPQNGRGALFAGGLAAILASTCCLGPLVLVALGFSGAWIGNLTVLEPYRPIFIGAALVALFFAWRRIYRPAQACKPGE VCAIPQVRTTYKLIFWFVAVLVLVALGFPYVMPFFY |
| 2709 |
PF07444 (Ycf66_N) |
 Estimated TM-score=0.71; hard target. |
>Ycf66 protein N-terminus (Length=75) ILGIFLAVAGAGLYFLRSVRPELSRDHDIFFAAVGLLCGLILLFQGWRLDPILQFGQFLLTGSAIFFAVETIRLR |
| 2710 |
PF03125 (Sre) |
 Estimated TM-score=0.71; easy target. |
>C. elegans Sre G protein-coupled chemoreceptor (Length=333) IRSQNTTSYFWLPLFFYDEPYWAQVVHSFIEIFLYIFVGYVVSVSIRIMLKVRLFHDNLVYIGTPIFALWSTLISAKLIV IAYRLKFLKVDYEIGEHITLWTDNPDKMLTVNSTEGLEPLMLGGFWVWHFGFSVMFGSFAVVVERVIASMLIDNYELSTD LYIPIILNTVYQLSTISISIALVFNKLGEIVLNASWITCFTISLSMYFYIKRINKRWLQEMEDPNRKRVFTVSQRFQVRE NLRALAFGKRIVFTVMGSLIFCGIGMVALSHDIVPTFLLHIGENLLHPLFVCVVTMYGHPSWKSRFKRSFPRIHFLRKPP NRVVSVEIMEDNV |
| 2711 |
PF14493 (HTH_40) |
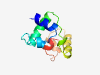 Estimated TM-score=0.71; trivial target. |
>Helix-turn-helix domain (Length=88) THRETLALCQQGKSVSEIALARNLSPSTIYGHLERLLQAGEAIDINTLVPQDHQQPIRQAIQALNTDKLKPVYEYLGGVY PYEEIRLV |
| 2712 |
PF13250 (DUF4041) |
 Estimated TM-score=0.71; hard target. |
>Domain of unknown function (DUF4041) (Length=56) KLVLRAFNGECDAAVSKVKYNNIQTMENRIQKAYDSLNKLSQTTHCEITPGYLNLK |
| 2713 |
PF05533 (Peptidase_C42) |
 Estimated TM-score=0.71; very hard target. |
>Beet yellows virus-type papain-like endopeptidase C42 (Length=86) YPDGRCYLAHMRYLCAFYCRPFRESDYALGMWPTVARLRACVEKNFGVEACGIALRGYYTSRNVYHCDYDSAYVKYFRNL SGRIGG |
| 2714 |
PF07351 (DUF1480) |
 Estimated TM-score=0.71; very hard target. |
>Protein of unknown function (DUF1480) (Length=79) MNQTKLRISSYEIDDAILSSPSQEDDITISIPCNSDQELCMQLDGWDEHTSIPALLNDKHILLYRKHYDRQKHAWVMRV |
| 2715 |
PF18871 (HEPN_Toprim_N) |
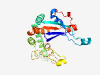 Estimated TM-score=0.71; very hard target. |
>HEPN/Toprim N-terminal domain 1 (Length=230) MGTEIQLKLGGLGLTYAKNHRGPDHGALFQPADRQRIRSDQVDYEYCEEEGEDPSPMEWAFARKLKHVVPRLQLLGFTID LAREAYSNWVVDWKEERESLQDDDDPPIPEPLSFDEFCAFATRQSILDLDDTFLSGDEEWRDRRKPKGRFASDMAIIERI PIGFDHDDMAWSERSYFGSRVNILHPYLMLQILAGNAANLETDVTWQYGPLVENGYTAAAAFTPSARRTE |
| 2716 |
PF05753 (TRAP_beta) |
 Estimated TM-score=0.71; hard target. |
>Translocon-associated protein beta (TRAPB) (Length=172) FALSQLSVANEDNSARLLVSKQILNKYLVENRDIVVKYTLYNVGSGAAVNVQLVDNGFHPEAFTVVGGQLSATIDRIAPQ TNVTHIAVVRPKAYGYFNFTAAEVNYQPAEEVTELQFAVSSEPGEGAIVAEAQFNKQFSSHYLDWFAFAIMTLPSLAIPY GLWFSSKSKYEL |
| 2717 |
PF05617 (Prolamin_like) |
 Estimated TM-score=0.71; easy target. |
>Prolamin-like (Length=65) ECWDSLMELKSCTGEVILFFLNGETHLGPNCCQAIRIIEHHCWPTMLSVLGFTPEEGDILRGYCD |
| 2718 |
PF09903 (DUF2130) |
 Estimated TM-score=0.71; hard target. |
>Uncharacterized protein conserved in bacteria (DUF2130) (Length=257) LKAKDDIIAYKDGEIERYKDMKARLSTKMVGESLEQHCETEFNKLRSTAFPRAYFEKDNDASEGSKGDFIFRECDEDGNE IVSIMFEMKNESDDSSHRHRNEDFFKKLDSDRTKKGCEYAVLVTLLEPESELYNQGIVDVSWRYDKMYVIRPQFFIPLIS ILRNAALNSLAYKAELAVVRNQNIDITHFEEQMEAFKNGFARNYDLASRKFQTAIDEIDKTITHLQKTKEALLSSENNLR LANNKAQDLTIKRLTRN |
| 2719 |
PF15257 (DUF4590) |
 Estimated TM-score=0.71; hard target. |
>Domain of unknown function (DUF4590) (Length=107) KDIDLRHEVKVFQQHCEGENICVYKGKLREGEFFQFISRRSRGFPFGLTFYLNGLQVEHLNSCCEFRHQKGSEPKGRDRH FSFVSMDGASPCYKCVLAFGLYKKSIR |
| 2720 |
PF06760 (DUF1221) |
 Estimated TM-score=0.71; trivial target. |
>Protein of unknown function (DUF1221) (Length=213) RDNIQINQRQCILLLDIFYSAYKSIADEMKDNLKFEERNLKWKVLEMPLRELHRIFKEGEAYIKESLESKDWWVKAITLY QNTDCVELHISNLLSCIPVVIEAIESAAQLSGWEQDEMQKKKRVYSNKYHKEWIDPELFQWKFAKQYLVTQDFCNRIDNV WKEDRWILKNKIQEKENSRFRKQERKLADLLLRNLDSSKSLKDKLLPSSILLG |
| 2721 |
PF17883 (MBG) |
 Estimated TM-score=0.71; easy target. |
>MBG domain (Length=99) QTYTGDSANTVIPSGITHNIQKSNDLLTWPTGAENVTLDSSDFSFASTNGNILTDKPVNVGTYHIVLNKQGLDKFKNLDS NFTWKYDPKTSYVVYKITK |
| 2722 |
PF12887 (SICA_alpha) |
 Estimated TM-score=0.71; very hard target. |
>SICA extracellular alpha domain (Length=179) NLLVQWIKAHSMKDMEKFGDQIWQDLDKLFKDMMENTEDSTETENTFCGMEDSKGVALGNSTEKELCKILLRIFFWINGL KLKWDNVAGWQWVEKEWKKESADEKQLQAYLRCLVGKVTMMRMFGTHCALDKVANLVKEAVDGYVGSFGSGDHYKVCEEV DISSVRMGGRLIWKELGQW |
| 2723 |
PF19138 (DUF5821) |
 Estimated TM-score=0.71; very hard target. |
>Family of unknown function (DUF5821) (Length=217) VRMLADERTLKDVMDDFIVASNAADLIHQDALELRTIEESPENSLLVTEDEVVALVRAGNRVGGLSTDDEEFVTTAYDAY MTEWEDAEAFTLRTPPISRVRETLESDISPEAEEDFDNILASLETARGDGDGLDEVTISLLVAAKNEALLYDISKWGEDV GIASKATFSRTKTKLEDMGLIDTEKVPIDVGRPRLRLKIGDDRLRNADNGQLATVAQ |
| 2724 |
PF10203 (Pet191_N) |
 Estimated TM-score=0.71; easy target. |
>Cytochrome c oxidase assembly protein PET191 (Length=67) SSCQDIREALAQCLQESDCIMVQRHTPRECLTSPLAEQLPMKCQQLRKGFAECKRGMVDMRKRFRGN |
| 2725 |
PF16438 (DUF5035) |
 Estimated TM-score=0.71; hard target. |
>Domain of unknown function (DUF5035) (Length=147) CSNSGESPVIKIEKLYVKAEPDSDSAKEEDYANKMDELPALKVGDEVKALLLLDGNGAELKTFKLQKDNDEALDTKLNYE KNEVSTEGNLTDEENGQLRFKDGVTQTKVLVQATIKQVDKNGDVRLEFYLSSKAECEGAQEEIGLKT |
| 2726 |
PF18474 (DUF5614) |
 Estimated TM-score=0.71; very hard target. |
>Family of unknown function (DUF5614) (Length=221) MASYAMLQERIAVAKDLLERVESISGGQKKGVEGKAKLCSKLRAELKFLQKVEAGEVVIKESHLQSTNLTHLKAIVESAE SLEDVVSVLHVFTYEDQAGEKQTLVVDVVANGGHTWVKAIGRKAEALHNIWQGRGQYGDKSIVKQAENFLQASRQQPVQY SNPHIIFAFYNGVSSPMADRLREMGISVRGDIVAVNSVAGAEEGEDALSDSSCESDADGDE |
| 2727 |
PF10713 (DUF2509) |
 Estimated TM-score=0.71; very hard target. |
>Protein of unknown function (DUF2509) (Length=131) MLMALSLMMLKALHYQQESAMLMMMDEKKYLNAFQRAESSLEWGKVQSWQFNMQEKENWFCQQQPEFHLQSCLKHYADNL FLLKGVAQFASGETLELYQWMKQMKKANRDTLIPIESGWLDFCPVPQAEFC |
| 2728 |
PF09991 (DUF2232) |
 Estimated TM-score=0.71; hard target. |
>Predicted membrane protein (DUF2232) (Length=287) AIYAILLLISMYVPILGTVVTFALPLPFILLTLRHRLSNVIVIFVAALLITVIVGQPLSLIKTVMFGLIGVVLGYMYKKE KKPVEILIAGTLAYLIGILLIYVASIKFFNIDLMKQMQNMFSESMAQSEKIVNAAGMPVSKEQKELFAQMSDILQTLFPS ILVLVSVCFSWITLILSGSVLRKLKYDVTPWPKFKDIQLPKSIIWYYVICILLATFIKVEPTSYLHMVFSNLNVIFALLL VLQGLTFITFLAHRKGFTKGVPIISFIACMFIPMLFPLVTILGIIDL |
| 2729 |
PF05677 (DUF818) |
 Estimated TM-score=0.71; easy target. |
>Chlamydia CHLPS protein (DUF818) (Length=361) AILDMHPKPSIAMFSSEQARTSWEKRQAHPYLYRLLEIIWGVVKFLLGLIFFIPLGLFWVLQKICQNFILPGAGGWIFRP ICRDSNLLRQAYAARLFSASFQDHVSSVRRVCLQYDEVFIDGLELRLPNAKPDRWMLISNGNSDCLEYRTVLQGEKDWIF RIAEESQSNILIFNYPGVMKSQGNITRNNVVKSYQACVRYLRDEPAGPQARQIVAYGYSLGASVQAEALSKEITDGSDSV RWFVVKDRGARSTGAVAKQFIGSLGVWLANLTHWNINSEKRSKDLHCPELFIYGKDSQGNLIGDGLFKKETCFAAPFLDP KNLEECSGKKIPVAQTGLRHDHILSDDVIKEVAGHIQRHFD |
| 2730 |
PF01785 (Closter_coat) |
 Estimated TM-score=0.71; easy target. |
>Closterovirus coat protein (Length=187) KNENYSSVDSSRLSDSEVKEVLEKSKESFKSELASTDEHFVYHIIFFLIRCAKISTSEKVKYVGSHTYVVDGKTYTVLDA WVFNMMKSLTKKYKRVNGLRAFCCACEDLYLTVAPIMSERFKTKAVGMKGLPVGKEYLGADFLSGTSKLMSDHDRAVSIV AAKNAVDRSAFTGGERKIVSLYDLGRY |
| 2731 |
PF09935 (DUF2167) |
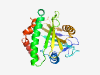 Estimated TM-score=0.71; hard target. |
>Protein of unknown function (DUF2167) (Length=239) PQQGEIKLPNGVATLQVPENFYYLSPEDTETVLVKIWGNPPGGEKTLGMIFPTRVTPYDNTAWGVTVEYVEDGYVSDEDA DKINYDELLSDMKESTANASQERIKQGYDAISLIGWAAKPYYDEKTHKLHWAKEIKFGSQEINTLNYNIRVLGRKGVLVL NFIAGMDQLDMINSKIDTVLKIADFNEGSRYADFNPDIDTVAAYGIGALVAGKVAAKVGILATILLFLKKLWIFLLIGA |
| 2732 |
PF06645 (SPC12) |
 Estimated TM-score=0.71; hard target. |
>Microsomal signal peptidase 12 kDa subunit (SPC12) (Length=70) IDFEGQRLAENLSTGLLSIVGVVAFLVGYITQNITYTLWIGLAGTALTFLVVVPPWPFFNRHPVKWLPST |
| 2733 |
PF17340 (DUF5370) |
 Estimated TM-score=0.71; hard target. |
>Family of unknown function (DUF5370) (Length=62) MGAIERSGYRFEPEFSVINQNGAVHVYQNGEFIEEIKFTFSGKFPEHDQIEELVDKYCEQHQ |
| 2734 |
PF05512 (AWPM-19) |
 Estimated TM-score=0.71; hard target. |
>AWPM-19-like family (Length=141) LLNFCMYVIVLGIGGWAMNRAIDHGFIIGPGFDLPAHFSPIYFPMGNAATGFFVTFALIAGVVGAASAISGINHIRSWTA DSLPSAAAAATVAWTLTLLAMGFACKEIELRIRNARLRTLEAFLIILSATQLLYIAAIHSA |
| 2735 |
PF12018 (FAP206) |
 Estimated TM-score=0.71; very hard target. |
>Domain of unknown function (Length=278) ILNEAIPATTQHIDIQLQTSQEQAYRYTAILEKASENPLKMKDLEPFMIKEALFNLRQYEVFLNIILSDIITCAQEVEMM IKQLAAQMEQLKMTVQAKTAVPTSQVYPVFIALSGLWSSFQDETVLLSVLSNLTTHLGPFLGSHEELFSEQIIQNYLEEV EIKTDVFRIKESMGTRVNVGDFKKVDWLFPETTANFDKLPIQYRGFCGYTLAVTDGLLLPGNPTIGILKHKDRYYTFNSK EAAYAFADNPNKYIEMIGEKAKKSAELIQLLELHQQFE |
| 2736 |
PF18757 (Nmad5) |
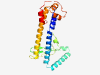 Estimated TM-score=0.71; hard target. |
>Nucleotide modification associated domain 5 (Length=202) ILTKDIKSLIVTNALTKAGIFEKEDKLFQDRADWAEKIRIEAIGGKNMEQGIQDIMSEIEKLTTKLPESLRTNITPINRS NCVGINLAGSRVYAYFNGARERFKYPDSRKHITKITPDNTTLLADNPLVNEFYELEKRHDELKSQRADITNNVEAVLLKV RSVKRLLEEWPEAAELLPKDMGKAAPVPAVRREALNTLIGLP |
| 2737 |
PF17583 (DUF5484) |
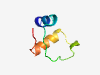 Estimated TM-score=0.71; hard target. |
>Family of unknown function (DUF5484) (Length=43) MNIKRMLFKQGLYTLNVTPKGDTTKWSVNDWIKFIDENGNWEI |
| 2738 |
PF14182 (YgaB) |
 Estimated TM-score=0.71; hard target. |
>YgaB-like protein (Length=76) EFEKLVSEQMKTMDKLLDLQSELDRCKQIEAELRHLERDARLRGIQNEIAVKRKHLADIQDMFQKQTEQVIRSYRS |
| 2739 |
PF18823 (InPase) |
 Estimated TM-score=0.71; easy target. |
>Inorganic Pyrophosphatase (Length=165) PTDAQKAAGNYKMGHATLHGLDITIENPRGSTRSGTDQDGKPWSVDMANHYGYIKRTEGADGDHVDAFIGPNPDSTHVFV VDQVDPRIGKFDEHKVMLGFDSLKAAREGYHANYEKGWKGAAAITPMSVDEFKAWLKNGDTTKPIDKKAGRSRTEKQAKA ARAAS |
| 2740 |
PF17803 (Cadherin_4) |
 Estimated TM-score=0.71; easy target. |
>Bacterial cadherin-like domain (Length=67) TITIDVVAAETPPTVNNDFFTTPEDIPVLGSVLSNDSDPNDETLTTTLLSEPSNGSVTLEQNGVFNY |
| 2741 |
PF07756 (DUF1612) |
 Estimated TM-score=0.71; easy target. |
>Protein of unknown function (DUF1612) (Length=127) FIYDLDWDEDERLGEWRTVLRELDSFPPVLQAIVALDAWNTLEVLQHAPWLGRLLAASILRQAGLTTAAHLAAFNLGLKS IPVDRRRHRDRETRLLAILAGLIAAAELGLKEHDRLTLARHSMERRL |
| 2742 |
PF12614 (RRF_GI) |
 Estimated TM-score=0.71; very hard target. |
>Ribosome recycling factor (Length=125) EDITIPLPSLIHRIGGEKAKQAKIIALQYDCELKRVRRSRNWRVIGEAVNIQSFTEQLKAQSDDNFRYLIRKIETALLGH ADKLEPLDEKLVRLIKQTPGITLGELIQLTQCTLTEARVARFNVD |
| 2743 |
PF16270 (DUF4923) |
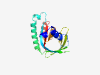 Estimated TM-score=0.71; hard target. |
>Lipocalin-like domain (DUF4923) (Length=173) LKDLFNKENVEKVVSAVTGKNTVDMTGTWSYTGAAIEFESDNLLMKAGGAVAATTAEAKLNEQLSKVGIKPGQMSFTFNA DSTFNAKLGTKTLNGTYMYNATEKHVTMKFVRLINMNAKVNCTSGSMDLLFESDKLLKLITFLSSKSSNATLKTISSLAN SYDGMMLGFSLEK |
| 2744 |
PF08560 (DUF1757) |
 Estimated TM-score=0.71; hard target. |
>Protein of unknown function (DUF1757) (Length=141) WFKNFAGIRQSDFEMLRVPNPSVEFCIHVTMRSVQTGALLGSVLGPLSAMLFEAKHMNSKKLTDKFVSGGTNGALIGAVM GPLLTYLALRDMNTVRLYDKCYRLRFDKQQLWQDRTCIVSAALGYLSSGSLGFVVGLDLAT |
| 2745 |
PF18647 (Fungal_lectin_2) |
 Estimated TM-score=0.71; easy target. |
>Alpha-galactosyl-binding fungal lectin (Length=99) ENRKAFASWERLNKKYRDFCEDVEPPQDTINWKWEKTYDKGTPDETQFVVHLSNEASAFDRSQCFESFDLILNSCDGNDP ENPMNWKFGGKYVRGSYRY |
| 2746 |
PF04065 (Not3) |
 Estimated TM-score=0.71; hard target. |
>Not1 N-terminal domain, CCR4-Not complex component (Length=230) DKRKLQGEIDRCLKKVAEGVEQFEDIWQKLHNAANANQKEKYEADLKKEIKKLQRLRDQIKTWVASNEIKDKRQLVENRK LIETQMERFKVVERETKTKAYSKEGLGLAQKVDPAQREKEETGQWLTNTIDTLNMQVDQFESEVESLSVQTRKKKGDKEK QDRIDELKRLIERHRFHIRMLETILRMLDNDSVPVDAIQKIKDDVEYYIDSSQDPDFEENEFLYDDLDLE |
| 2747 |
PF14761 (HPS3_N) |
 Estimated TM-score=0.71; easy target. |
>Hermansky-Pudlak syndrome 3 (Length=223) RVITVHNFLGQNVTQIEEPTATCTATSQRREMLLLALSTHCVEVWELMNSDVKLRTVFPTVDMINQMVHCAKGDYIVTLE SKYLRDTVSNNHANKVTSNFVRIYVNWATVKDQSQPMRARIAGRVTPSLNRPLNSLEMIELPLNMQPTLIACCQSTGNLL VASGNTAVLHEFKIETQQVSKMKFIDFESRPWSLGFSFAPTHMEIVEDFIAIMDNVNLSVFRL |
| 2748 |
PF06664 (MIG-14_Wnt-bd) |
 Estimated TM-score=0.71; hard target. |
>Wnt-binding factor required for Wnt secretion (Length=299) KCDEIIVLHLGYLNYTQYQINISFHNLSDLKYKIKEVNFTWKMYNSSFSQVEIWFRFVFVVLTFMVTCLFAHSLRKFSMR DWGIEQKWMSILLPLLLLYNDPFFPLSFLVNSWFPGMLDDLFQSLFLCALLLFWLCVYHGIRVQGERKCLTFYMPKLFIV GLLWLSAVTLGIWQTVNELHDPTYQYKVDTGNFQGMKIFFVVVAALYILYLVFLIVRACSELKNMPYVDLRLKFLTALTF VILIISIVILYLRFGSKVLQDNFVAELSTHYQNSAEFLSFYGLLNFYLYTLAFVYSPSK |
| 2749 |
PF13284 (DUF4072) |
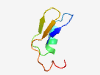 Estimated TM-score=0.71; hard target. |
>Domain of unknown function (DUF4072) (Length=47) LVIQSVAPLSDAHHKPLLALARGARIVQIDAHALRIEDANPAQRPDI |
| 2750 |
PF11817 (Foie-gras_1) |
 Estimated TM-score=0.71; easy target. |
>Foie gras liver health family 1 (Length=268) RFNHARLLADSLAIRIIRCLLWAGQTAAAVRSWVSHRDRTQDIVNRRGKGSRNYGWEAWESRWSMVMAELIRRAEISPQE QYLAPASHEKPTFTGDGLYPWEQLHHEGYWLYRSAKHAIKRRALAQQIPDEDRPPPGQSPASQIASKSYLYDTYLAPETH VEAPLSGSKGFDHSSLILNTLKAALEEFAKRNQTRKVESLSLEMAEEYVRIGSWSDGLNLLRPLWSTLSWRGSGWWPLME RFGLALRECALQIGDGEMVVRIDWELLS |
| 2751 |
PF06763 (Minor_tail_Z) |
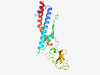 Estimated TM-score=0.71; very hard target. |
>Prophage minor tail protein Z (GPZ) (Length=192) MKGLENAIRNLNSLDRQMVPRASIWALNRVAQKAVSVATRKVARETVAGDNQVRGIPLKLVRQRVRLFKAGTDGKRSARI RINRGNLPAIKLGAAQVRMSKRRGKLLYRGSVLKIGPYLFRDAFIQQLANGRWHVMRRVNGKNRYPIDVVKIPLSGPLTQ AFESATQSLIDEEMPKQLGYALKQQLRLYLSR |
| 2752 |
PF06649 (DUF1161) |
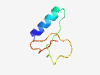 Estimated TM-score=0.71; hard target. |
>Protein of unknown function (DUF1161) (Length=47) CGELKDEIAAKLTAAGISPSVLAIVDKDWVGAGKIVGSCENGTKRIV |
| 2753 |
PF16080 (Phage_holin_2_3) |
 Estimated TM-score=0.71; hard target. |
>Bacteriophage holin family HP1 (Length=55) DKYTSPTAYTWGAFTAMLGALSLNDWAIVIGIICTIGTFAVNWYYKHREFNRHEK |
| 2754 |
PF10330 (Stb3) |
 Estimated TM-score=0.71; hard target. |
>Putative Sin3 binding protein (Length=89) ITPAMLAKYHLPEILLQHGPLAIRHVMGYLTTSVPGFSGIPPAKARRLVVAALEGRGSDDKSDVVYEKVGWGRWDARRRG EPSRDSSRH |
| 2755 |
PF14463 (E1-N) |
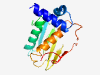 Estimated TM-score=0.71; hard target. |
>E1 N-terminal domain (Length=150) MNKATQQNAMMLASLLGVGEAEAGERLARTVLITAAPGWKSGWAVEVGELIGRTVQVSHQQEPTDPDLELVIGDVTPRTS ARRVYADLGSEGAAASLEPVAKLAGEPHGLYAAAAACAVSAVVVHAVIDAADLPQARLPMRLDYAQLGVP |
| 2756 |
PF06301 (Lambda_Kil) |
 Estimated TM-score=0.71; easy target. |
>Bacteriophage lambda Kil protein (Length=42) NHQALMAAQSKAVIARFLGDAGMWLQANQQMKQAVSMPWYRR |
| 2757 |
PF11597 (Med13_N) |
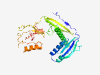 Estimated TM-score=0.71; hard target. |
>Mediator complex subunit 13 N-terminal (Length=286) KASNILTHYYKLAHVGSITYTIYASSENNDQALLELELTIRNKYPEILITYYNKNLYYFAFGHNLINSDSPIDLSTEFHQ LSCKSSESVTADQLANPIKNHANNNENLTYAGLSFLKAVKKMILYNLSLNGSIRLFGNYCVSPNDDQSYSILCIDPVLFQ NGDLLVSCLEKPNVRLFSSMVAYPEELAIETNFVIYLIPSGIRCHLFDPTNLRNNFIENPQVENEKLPEMLRLTTGIDCD KSKLWVKLIPNLKHLNNQTSFVGKFIHSVDNKKFILWPWDLCLLQF |
| 2758 |
PF11115 (DUF2623) |
 Estimated TM-score=0.71; hard target. |
>Protein of unknown function (DUF2623) (Length=92) MNNHFGKGLMAGLKATHADSAVNVTKFCADYKRGFVLGYSHRMYEKTGDRQLSAWEAGILTRRYGLDKDMVMDFFRENNS CSTLRFFMAGYR |
| 2759 |
PF16149 (DUF4857) |
 Estimated TM-score=0.71; very hard target. |
>Domain of unknown function (DUF4857) (Length=270) FSMYSTIIDDFVSMGHEEGKGMMRRDQSGNVYTQEQVDSILPFFYIRQLMADERFPDTIKGIAVTPREIQLTNFSFRSAP SDINASRIELYPLLESMSGRVDLAMPDDVFRITNKGIEFIKMKTNTVDADKSSQFTEAMVKKGFKFPAQRLAGNPTTRKE YDEGYLILDNEGKLFHLKQTKGRPYVRAISLPEGVKAKHLFITEFKSRKTLGFLTDMDNNFYVLNNKTYDLVKTGVPAFN PEQDALSIFGNMFDWTVCIKTGKADEYYAL |
| 2760 |
PF17400 (DUF5406) |
 Estimated TM-score=0.71; very hard target. |
>Family of unknown function (DUF5406) (Length=113) SFDPNYIHCFSKKTIKLTFQQWEYTGTAFVEIGGNCTFADMLSEFENGDTLLSLLKQKTSKLNFDLKDLGQDEEGKNWFR AILISDTGEKCETEDFLDALPEMLVGIDLIDIQ |
| 2761 |
PF10563 (CA_like) |
 Estimated TM-score=0.71; easy target. |
>Putative carbonic anhydrase (Length=178) TEMNLCNIHFHKNAEHKGGEFTLYAGNGDGEGYHTGYRYSGKLSDAELAPVKNKVCPSKHGDLKPGDTIEVHYVHSTAQV EPGPTLGACLSDANKNPQLRVETQVYVLVNDKNAYDFEELTEHEVQDGVHQAVAIPTNTGKPIQYMGSTTGPGYNEKGSP FQVTWSVRPKVAKVNIES |
| 2762 |
PF12094 (DUF3570) |
 Estimated TM-score=0.71; easy target. |
>Protein of unknown function (DUF3570) (Length=422) ASLAAAACALLNSAVQAQENTQANEWQFDTAILYYGETDRVQLGEGVFNAKKDFGDEHIFNGKIAIDTLTGASASGAVAQ QGVQTFTRPSGNGQYVVNAGETPLDDTFKDTRVQLSAQWTQPLWENITGSTGVHLSKEYDYLSLGVNGSLARDFNQKNTT LSAGVSYQYDSIDPVGEAPTGLSPMVVNNGQFSDESAFETAFNATRATDSKDKNTVDLLLGVTQVINRRMLMQFNYSYSI ADGYLTDPYKLLSVVNTEGVTQDILHENRPDKRTKQSFYWQTKYASDYGIADVSYRYATDDWDIDSHTLDSRFRVKLSDN SYIQPHFRYYQQSAADFYQPFLIQGSLLPSVASADYRLGEMTAYTIGLKYGQALDNGREWAVRLEYYQQSPKNAGFEEPG ALQGLDLYPSIDAIIAQVSYSF |
| 2763 |
PF12274 (DUF3615) |
 Estimated TM-score=0.71; easy target. |
>Protein of unknown function (DUF3615) (Length=94) YNDDHNLLEDHAFELKGVINYEPIIESRRWFDHINFTATTKGLNGLDSDHLFFAEAMSLKGEKDYVVTCCSLISSDDNGN CYTCKIGNKSMKHP |
| 2764 |
PF10520 (TMEM189_B_dmain) |
 Estimated TM-score=0.71; easy target. |
>B domain of TMEM189, localisation domain (Length=178) IVTADFASGIVHWGADTWGSVDIPVIGKAFIRPFREHHIDPTAITRHDFIETNGDNCMLSILPLAHMAYKFLTYQPDAVA ETYPWECFVFALAVFVTLTNQIHKWSHSYFGLPRWVTLLQDCHLVLPRKHHRIHHVSPHETYYCITTGWLNYPLDKVGFW RKMERLIESVTGQKPRSD |
| 2765 |
PF09838 (DUF2065) |
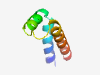 Estimated TM-score=0.71; hard target. |
>Uncharacterized protein conserved in bacteria (DUF2065) (Length=52) IAAVGVMMLLEGLPLLLAPEQWRRYVNYILTLAPGQLRFIGLAMLLLAVVLV |
| 2766 |
PF11948 (DUF3465) |
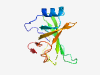 Estimated TM-score=0.71; easy target. |
>Protein of unknown function (DUF3465) (Length=122) VLLCFGVSHVQANDARLKQAYQNHQSDVQVRGSGLVSRLLPDDNKGSRHQKFILRLDSGQTLLVAHNIDLAPRIQGLRKG DRVEFYGEYEWNKKGGVMHWTHKDPRNHHAHGWLKHNGKIYE |
| 2767 |
PF10069 (DICT) |
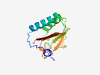 Estimated TM-score=0.71; very hard target. |
>Sensory domain in DIguanylate Cyclases and Two-component system (Length=137) DLSVYQLVMGVQVPPKPLSLSPATLLSLVRAQIDLLIEQQIAATLWVKLPPEKIWQSELARYQSSVGASSIIYTCQIDEN EKQGAGEDDEAGEEKTSSSFSSTHHVTVHLTPDSQLRRENFLMVLSPQFCSLILAHR |
| 2768 |
PF16288 (DUF4934) |
 Estimated TM-score=0.71; trivial target. |
>Domain of unknown function (DUF4934) (Length=102) LKVSDLGKTIRYIPLETTDSCLIGDFPNIKLLDDKIMVYNGKQCLLFDKETGKFICSVGHRGDDPEAYSSTCGYLNPQNQ LLYFNRDPNQLVKYNQKGNFAG |
| 2769 |
PF10774 (DUF4226) |
 Estimated TM-score=0.71; hard target. |
>Domain of unknown function (DUF4226) (Length=112) QAGSSVAVIQERQALLARQHDAVAEADRELADVLASAHAAMRESVRRLDAIAAEIDRAVPDQDQLAVDTPMGAREFQTFL VAKQREIVAVVAAAHELDRAKSAVLKRLRAQY |
| 2770 |
PF10561 (UPF0565) |
 Estimated TM-score=0.71; hard target. |
>Uncharacterised protein family UPF0565 (Length=305) RINGVKGHQNRTNSIIYCPPLLRTNPQKDKCSAIIYFGGDVQDIPEKMESNRDNKNYIKWNLENTALLLRESFPQSHIIV VRPMRMEYSTFSCFDNFVRGNNAGIPDHTPMHFSLQHLEELLINLTKKLTKPILDQEFLHKLLAASSSKTLAGDVSCKQN NVDILQSDIYNSTSGDSNEGDLTDLLWWRENLNLDKASLKLIGFSKGCVVLNQFIYEFHYYKTLTPDDSTMMRLVSRITD MYWLDGGHGGGKNTWITSRSLLETLCRLCINVHVHVTPYQIQDDHRPWIRKEEKAFTDLLKRLGA |
| 2771 |
PF03896 (TRAP_alpha) |
 Estimated TM-score=0.71; hard target. |
>Translocon-associated protein (TRAP), alpha subunit (Length=286) KLLIFALLVLPAVLLTVDSGSRLMAHATEDELDEELDVEVESDDASVTQTDEAETDDEPETTKSPDADTFLLFTRPLHSS GSQLELPAGYPVEFLVGFANKGSDDFIVETVEASFRYPMDFNYYIQNFSAIPYNREIKPGHEATVSYSFLPSESFAGRPF GLNIALNYRDASGNQFSEAVFNETVSITEVDEGLDGETFFLYVFLAAVVVLLLVLGQQFLGSYGKRKRSPAVRKTVETGT TNTKDVDYDWIPAETLKKIQNSPKGGKVSPKQSPRQRKAKRAAGSD |
| 2772 |
PF01696 (Adeno_E1B_55K) |
 Estimated TM-score=0.71; easy target. |
>Adenovirus EB1 55K protein / large t-antigen (Length=382) GNNSRTELALSLMSRRRPETVWWHEVQSEGRDEVSILQEKYSLEQLKTCWLEPEDDWEVAIRNYAKISLRPDKQYRITKK INIRNACYISGNGAEVIIDTQDKAAFRCCMMGMWPGVVGMEAVTFMNIRFKGDGYNGIVFMANTKLILHGCSFFGFNNTC VEAWGQVSVRGCSFYACWIATSGRVKSQLSVKKCMFERCNLGILNEGEARVRHCAATETGCFILIKGNASVKHNMICGHS NERPYQMLTCAGGHCNILATVHIVSHARKKWPVFEHNVITKCTMHIGGRRGMFMPYQCNMNHVKVMLEPDAFSRVSLTGI FDMNIQLWKILRYDDTKPRVRACECGGKHARFQPVCVDVTEDLRPDHLVLACTGAEFGSSGE |
| 2773 |
PF06023 (Csa1) |
 Estimated TM-score=0.71; easy target. |
>CRISPR-associated exonuclease Csa1 (Length=290) MFFTHSDMLLLSKRIKKLPKNVDEELRGWNWSEPPVYTRSLSQVSISEMVYCSTLRNVYLKVKGFRGEIGRQILQGSLIH TIYAIGIEAIKRFIYSRESIDGSTLRTLMGDEFYSLLKDLREEEGIYAKVLWDHITNIYSAELDRVRSKFTNLTRDSLVS QVVPFYVEFPVDGSLLGLTNLRVDAFIPHLPLIAEMKTGKYRYTHELSLAGYALAIESQYEIPIDFGYLCYVTVTEKEVK NNCKLIPISDSLRSEFLDMRDKAQDIMDKGVDPGIAKDCESDCMFYKVCH |
| 2774 |
PF10048 (DUF2282) |
 Estimated TM-score=0.71; very hard target. |
>Predicted integral membrane protein (DUF2282) (Length=49) WEKCAGIVKVGMNACGTSKHGCGGAAKTDADPEEWIMVPQGTCDKIVGG |
| 2775 |
PF18832 (LPD18) |
 Estimated TM-score=0.71; hard target. |
>Large polyvalent protein-associated domain 18 (Length=83) SGTLVSLDTTDFHIKGKEGKWLAFDSIIIEGRQFFLMEHETYGKEVAWVVLDEEGKLVVDHAYHGFDQSVQQQIKDYLNP PQL |
| 2776 |
PF10789 (Phage_RpbA) |
 Estimated TM-score=0.71; hard target. |
>Phage RNA polymerase binding, RpbA (Length=110) ADLKVTSIRTDANPHNHNRVRKAWVLHCDDASAKKLQSLPQETRFMIYGFIDNDVSDMWIHLMRKHYKDSIEAGAKIVLD KDGSERLEDFYCVDADEQLIAAGEIVASKI |
| 2777 |
PF00660 (SRP1_TIP1) |
 Estimated TM-score=0.71; hard target. |
>Seripauperin and TIP1 family (Length=90) LAQTAAESAELSAILADLQAHESDYIGQIASGNAPPSGLLSLVQKVLTYTDDSYTTLYSNVNIDEVNSYITKLPWYSTRL SSEIYEATKN |
| 2778 |
PF04927 (SMP) |
 Estimated TM-score=0.71; hard target. |
>Seed maturation protein (Length=54) KYGDCFNVKGDLADQPIAPQDAAMMQSAEAMVFGKNQKGGPASVMQAAAAKNER |
| 2779 |
PF10801 (DUF2537) |
 Estimated TM-score=0.71; hard target. |
>Protein of unknown function (DUF2537) (Length=79) TPWGTGLTVAAFVAAVVGVAIVVLSRGLISIHPLLAVGLNLVAVGGLAPTVWGWRRKPVLRWFVLGSAVGVAVGWIVAL |
| 2780 |
PF11193 (DUF2812) |
 Estimated TM-score=0.71; hard target. |
>Protein of unknown function (DUF2812) (Length=105) DVQKTEQWLTDQAKAGFRVKEMHRFKRGFTFEKGQPKDVTYRIGYDKIKPATLSNTMRNDGWEKVAQSGKWYVIANERPQ AEVTTSTSRDAIIKRNNFIYYAFMA |
| 2781 |
PF14441 (OTT_1508_deam) |
 Estimated TM-score=0.71; easy target. |
>OTT_1508-like deaminase (Length=63) THAELLLIDYIEQHEEIRETNDKYIGCSKPACYLCHLYIRHHPGNYALPDTSNKLYVKWRIPD |
| 2782 |
PF16910 (VPS13_mid_rpt) |
 Estimated TM-score=0.71; hard target. |
>Repeating coiled region of VPS13 (Length=233) FEYKPLDNRADNAAALYMRNIDIIYNPEIIRELIVFFTPPETSADSINALIEVAGDTFEGIKKQTKASLEYALEQHTTFD LKVDMDAPVIIIPENCTSTSCRAIVIDAGHINVESNLSSPEMRTQMKSKSGAEMSAEDNLQFRSLMYDRFTIQLTETKVL VGDSLDTCLVQIRNPRPELSYLHLIDHIDMMFLLELCIIRKSVDMPRFKVSGHLPLLKVNFSDTKYKALMQLP |
| 2783 |
PF03312 (DUF272) |
 Estimated TM-score=0.71; very hard target. |
>Protein of unknown function (DUF272) (Length=125) YLWNLDSKTEGLFVSKNHSLAQGHHFEGIFKKNPDGRWTCQKYENPIEGLLTGGINPNNKIYFTVKIDKYQPAKGNTKYG HATAKYIGDVLEGDSENTKLSAECNGKMVNIQRRGIGDKDFVWMS |
| 2784 |
PF08896 (DUF1842) |
 Estimated TM-score=0.71; hard target. |
>Domain of unknown function (DUF1842) (Length=114) VGLFPVSYVVGTGLPGAPTLRLNLLVDTPRRKVVGNATITQAVSPPLDLHVDVWGPYTYMALMPPHGTRILVTVQGNSGG PTSNAMTTFRMHLVVGADWQAGIASYSYWSNGQW |
| 2785 |
PF01677 (Herpes_UL7) |
 Estimated TM-score=0.71; trivial target. |
>Herpesvirus UL7 like (Length=211) MVLEVNRCENLCVACNSPVLVRDGALCVSQTLDYVKQKLVSDTFMGFAMACLLDCEDLVDSINLAPHVFAQRVFVFRPPN SYLLEMCVLSSMLENCETYTRRFVESLVSRARFIYSKSPCIDACFLLHSIEMMASTIIDYFKLDPEGAQPRYPGLMMYKL HGAIEDGSFESKGLLRPIYHESFKLCSDPDASFDEEEETGEAGVTFNVFYC |
| 2786 |
PF06635 (NolV) |
 Estimated TM-score=0.71; easy target. |
>Nodulation protein NolV (Length=207) MTADISVAPAAPQMRPLGPLIPASELEIWDNAAKACAAAERHQQHVRSWARAAYQRELARGHTEGLNAGAEEMAALISQA VAEVARRKAVLEQQLPQLVLEILSELLGAFDPGELLVMAVRHAIERQYSGAEVCLHVYPTQVDMLAREFAGWDGQDGRPR VRIKPDPTLSPRRCVLWSEYGNVDLGLDAQMRALRLGFGSLSEKGEL |
| 2787 |
PF12304 (BCLP) |
 Estimated TM-score=0.71; hard target. |
>Beta-casein like protein (Length=184) MKTGLTLILIGHINFILGAIVHGSVLRHISKPSQHISTEYTVANIISVTSGLLSIATGIIAIVVSRNIHVLKLQIGLLIA SFLNALLSAACCTGLLMAISVTVAHSGQGLMQGCNDTEVPINARSPVSAQCPFDTTRIYDTTLALWFPCAVLSAVEVALS VWCFIVGLALRGLAPCGKSYIKEQ |
| 2788 |
PF15610 (PRTase_3) |
 Estimated TM-score=0.71; easy target. |
>PRTase ComF-like (Length=264) PYTYSLHKIHHSNNFGFSADDYSRFKFGDDRVAKKFGKDLADGFIKGFLEDNQISEQIVVISSPYCFIPTATFAMKNYFV YQLNHWLVENGRMVVQEAKVHRTITYKDDYGALSAEDRIKLIGNDSFHIDKDFLRDKVLFFLDDIKITGSHEKMILKMVK EYELKNDIYMLYFAELANHDIHPNIENHLNYHKVRSVFDLDEIIKSGNFCFNTRIVKYILNCDFNSFSIFLERQTEDFIY NLYNLSLGNSYHTIDCYSGNLNFL |
| 2789 |
PF11165 (DUF2949) |
 Estimated TM-score=0.71; easy target. |
>Protein of unknown function (DUF2949) (Length=56) YTRFINFLQEDLAISAAAIAVALRHREQDPGPLPMILWQYGLITLEQLDRIYDWLE |
| 2790 |
PF03752 (ALF) |
 Estimated TM-score=0.71; hard target. |
>Short repeats of unknown function (Length=44) RAEIQRLLADPSTGETVREAAQKALAAGTPAALRQFLATDLQSA |
| 2791 |
PF16430 (DUF5027) |
 Estimated TM-score=0.71; easy target. |
>Domain of unknown function (DUF5027) (Length=187) EDIVIDNESDIYNIGDEIQVKNVDYGTNREYVAKSYIVNSVKIYDTAAEIDGVQDKLIETDYYMGADPEKPAILKKDEVA CGKLLLCDISVKNIEDEICTVGDISLVYEINGACQLLGYPIYFSNAKDNEHGIYDYTLVQGQSLDAQIGFCVDPALLQID NMDLSKLYLSVNFNGDEENRQFIDLRL |
| 2792 |
PF14913 (DPCD) |
 Estimated TM-score=0.71; hard target. |
>DPCD protein family (Length=172) GRRKVHYLFPDGKEMAEEYEDETSELLVRKWRVKSALGALGQWQIEVGEPASHGAGNLGPELIKESNANPIFMRKDTKMS FQWRIRNLPYPKDVYTVCVDQKERCVVIRTSNKKYYKKFSIPDLDRYQLPLDDSLLSFAHANCTLIISYQKPKEVLVAES ELQKELKKVKTA |
| 2793 |
PF18738 (HEPN_DZIP3) |
 Estimated TM-score=0.71; hard target. |
>DZIP3/ hRUL138-like HEPN (Length=147) PGKLDTVLSSPPVHKALQLLKKKKVLNPSQWGKLYPAIKSSVSSQHFDITLLMVLLRNICGLVPPATGWNTLPPAADTTL EADIIRIKYYRDKVYGHAIEASVDDASFNQHWMDIRGALVRLGGARYQGDIDNLKKEGMDPEIEEHY |
| 2794 |
PF07865 (DUF1652) |
 Estimated TM-score=0.71; hard target. |
>Protein of unknown function (DUF1652) (Length=67) LELRRIVETSFLPLHCQCTVGHDGAMTVRICDPASGRVDLLVTGIATRGLTSSRALSELIAELRYDL |
| 2795 |
PF09527 (ATPase_gene1) |
 Estimated TM-score=0.71; easy target. |
>Putative F0F1-ATPase subunit Ca2+/Mg2+ transporter (Length=51) GNAATIGTHMVVSTFVGMGIGWYLDKWLGTKPWLLLVFLCFGIAAGFKNVY |
| 2796 |
PF14177 (YkyB) |
 Estimated TM-score=0.71; very hard target. |
>YkyB-like protein (Length=138) TVENLSKAVYTVNRHAKTAPNPKYLYLLKKRALQKLVKEGKGKKIGLHFSKNPRFSQQQSDVLISIGDYYFHMPPTKEDF EHLPHLGTLNQSYRNPKAQMSLTKAKHLLQEYVGMKEKPLVPNRRQPAYHKPVFKKLG |
| 2797 |
PF18476 (PIN_8) |
 Estimated TM-score=0.71; hard target. |
>PIN like domain (Length=229) FIFDTNVLLKLYSYQPNTLSDFFSILKKLGDKIWIPHQVGLEFQNRRLDIKAKEKQKFSELEKQLDEILKVENKINQLFL KGRFPNLDVATENLFNDIRKLVDGYKNTLNQCNESQPNIRSHDSIREKIDDCFKGKVGSPFSQEDLEKIYEDGEKRYENK IPPGFKDNNKNSSTDYFYKGVEYKRKFGDLILWKQIIEKAKDENIKNVIFITDDEKEDWWYIAKENGDK |
| 2798 |
PF11874 (DUF3394) |
 Estimated TM-score=0.71; hard target. |
>Domain of unknown function (DUF3394) (Length=179) GIVVFVVATIAMLIFAAATQGFMLTRNRWYESILLLLVAFTLFRPGFWMDMIHDPYRSIPASQFVQALGEMDDDSTLRVQ VAGEDAYGDPLTTYMTLPVPEGDTGEERLENFGLELLVKDGDLVVDMVAFGSQAEKLGFDFDQKIIEVMAPVDRWAKEWM WIPALAVFALIVLMQRRRR |
| 2799 |
PF05834 (Lycopene_cycl) |
 Estimated TM-score=0.71; easy target. |
>Lycopene cyclase protein (Length=398) DLVVIGCGPAGLALAAESAKLGLNVGLIGPDLPFTNNYGVWEDEFIGLGLEGCIEHVWRDTIVYLDENDPILIGRAYGRV SRDLLHEELLRRCVESGVTYLSSKVERITEATNGNSLIECEGDIIIPCRLATVASGAASGKLLQYELGGPRVCVQTAYGV EVEVESIPYDPSLMVFMDYRDFAKHSAESVEAQYPTFLYVMPMSPTKVFFEETCLASREAMPFNLLKTKLMSRLKTMGVK ITKVYEEEWSYIPVGGSLPNTEQKNLAFGAAASMVHPATGYSVVRSLSEAPNYAAAIAKILQQGRSKQMLDSGRYTTNIS KQAWETLWPLERKRQRAFFLFGLALIVQLDIEGTRTFFRTFFRLPTWMWWGFLGSSLSSTDLLLFALYMFIIAPQSLR |
| 2800 |
PF17436 (DUF5415) |
 Estimated TM-score=0.71; hard target. |
>Family of unknown function (DUF5415) (Length=66) MAKAKTPFEKAASKILERRGVSETQWKQEVLEKAQFDLLRGKDKDLEGYIYQREREKLVFDEIIKH |
[an error occurred while processing this directive]
zhanglab![]() zhanggroup.org
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218