


Go to page (17 pages in total): [First page] << < 3 4 5 6 7 8 9 10 11 12 13 > >> [Last page] [All entries in one page].
| # | Uniprot ID | D-I-TASSER-AF2 models ranked by estimated TM-score (click to view) |
Full name and sequence |
| 701 |
O95299 (NDUFA10) |
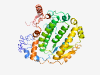 Estimated TM-score=0.925 |
>NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10, mitochondrial (Length=355) MALRLLKLAATSASARVVAAGAQRVRGIHSSVQCKLRYGMWHFLLGDKASKRLTERSRVI TVDGNICTGKGKLAKEIAEKLGFKHFPEAGIHYPDSTTGDGKPLATDYNGNCSLEKFYDD PRSNDGNSYRLQSWLYSSRLLQYSDALEHLLTTGQGVVLERSIFSDFVFLEAMYNQGFIR KQCVDHYNEVKSVTICDYLPPHLVIYIDVPVPEVQRRIQKKGDPHEMKITSAYLQDIENA YKKTFLPEMSEKCEVLQYSAREAQDSKKVVEDIEYLKFDKGPWLKQDNRTLYHLRLLVQD KFEVLNYTSIPIFLPEVTIGAHQTDRVLHQFRELPGRKYSPGYNTEVGDKWIWLK |
| 702 |
P13945 (ADRB3) |
 Estimated TM-score=0.925 |
>Beta-3 adrenergic receptor (Length=408) MAPWPHENSSLAPWPDLPTLAPNTANTSGLPGVPWEAALAGALLALAVLATVGGNLLVIV AIAWTPRLQTMTNVFVTSLAAADLVMGLLVVPPAATLALTGHWPLGATGCELWTSVDVLC VTASIETLCALAVDRYLAVTNPLRYGALVTKRCARTAVVLVWVVSAAVSFAPIMSQWWRV GADAEAQRCHSNPRCCAFASNMPYVLLSSSVSFYLPLLVMLFVYARVFVVATRQLRLLRG ELGRFPPEESPPAPSRSLAPAPVGTCAPPEGVPACGRRPARLLPLREHRALCTLGLIMGT FTLCWLPFFLANVLRALGGPSLVPGPAFLALNWLGYANSAFNPLIYCRSPDFRSAFRRLL CRCGRRLPPEPCAAARPALFPSGVPAARSSPAQPRLCQRLDGASWGVS |
| 703 |
Q9NX20 (MRPL16) |
 Estimated TM-score=0.925 |
>39S ribosomal protein L16, mitochondrial (Length=251) MWRLLARASAPLLRVPLSDSWALLPASAGVKTLLPVPSFEDVSIPEKPKLRFIERAPLVP KVRREPKNLSDIRGPSTEATEFTEGNFAILALGGGYLHWGHFEMMRLTINRSMDPKNMFA IWRVPAPFKPITRKSVGHRMGGGKGAIDHYVTPVKAGRLVVEMGGRCEFEEVQGFLDQVA HKLPFAAKAVSRGTLEKMRKDQEERERNNQNPWTFERIATANMLGIRKVLSPYDLTHKGK YWGKFYMPKRV |
| 704 |
Q6FI13 (H2AC18) |
 Estimated TM-score=0.925 |
>Histone H2A type 2-A (Length=130) MSGRGKQGGKARAKAKSRSSRAGLQFPVGRVHRLLRKGNYAERVGAGAPVYMAAVLEYLT AEILELAGNAARDNKKTRIIPRHLQLAIRNDEELNKLLGKVTIAQGGVLPNIQAVLLPKK TESHHKAKGK |
| 705 |
P45381 (ASPA) |
 Estimated TM-score=0.925 |
>Aspartoacylase (Length=313) MTSCHIAEEHIQKVAIFGGTHGNELTGVFLVKHWLENGAEIQRTGLEVKPFITNPRAVKK CTRYIDCDLNRIFDLENLGKKMSEDLPYEVRRAQEINHLFGPKDSEDSYDIIFDLHNTTS NMGCTLILEDSRNNFLIQMFHYIKTSLAPLPCYVYLIEHPSLKYATTRSIAKYPVGIEVG PQPQGVLRADILDQMRKMIKHALDFIHHFNEGKEFPPCAIEVYKIIEKVDYPRDENGEIA AIIHPNLQDQDWKPLHPGDPMFLTLDGKTIPLGGDCTVYPVFVNEAAYYEKKEAFAKTTK LTLNAKSIRCCLH |
| 706 |
Q96GA7 (SDSL) |
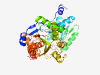 Estimated TM-score=0.925 |
>Serine dehydratase-like (Length=329) MDGPVAEHAKQEPFHVVTPLLESWALSQVAGMPVFLKCENVQPSGSFKIRGIGHFCQEMA KKGCRHLVCSSGGNAGIAAAYAARKLGIPATIVLPESTSLQVVQRLQGEGAEVQLTGKVW DEANLRAQELAKRDGWENVPPFDHPLIWKGHASLVQELKAVLRTPPGALVLAVGGGGLLA GVVAGLLEVGWQHVPIIAMETHGAHCFNAAITAGKLVTLPDITSVAKSLGAKTVAARALE CMQVCKIHSEVVEDTEAVSAVQQLLDDERMLVEPACGAALAAIYSGLLRRLQAEGCLPPS LTSVVVIVCGGNNINSRELQALKTHLGQV |
| 707 |
P25325 (MPST) |
 Estimated TM-score=0.925 |
>3-mercaptopyruvate sulfurtransferase (Length=297) MASPQLCRALVSAQWVAEALRAPRAGQPLQLLDASWYLPKLGRDARREFEERHIPGAAFF DIDQCSDRTSPYDHMLPGAEHFAEYAGRLGVGAATHVVIYDASDQGLYSAPRVWWMFRAF GHHAVSLLDGGLRHWLRQNLPLSSGKSQPAPAEFRAQLDPAFIKTYEDIKENLESRRFQV VDSRATGRFRGTEPEPRDGIEPGHIPGTVNIPFTDFLSQEGLEKSPEEIRHLFQEKKVDL SKPLVATCGSGVTACHVALGAYLCGKPDVPIYDGSWVEWYMRARPEDVISEGRGKTH |
| 708 |
Q9NP77 (SSU72) |
 Estimated TM-score=0.925 |
>RNA polymerase II subunit A C-terminal domain phosphatase SSU72 (Length=194) MPSSPLRVAVVCSSNQNRSMEAHNILSKRGFSVRSFGTGTHVKLPGPAPDKPNVYDFKTT YDQMYNDLLRKDKELYTQNGILHMLDRNKRIKPRPERFQNCKDLFDLILTCEERVYDQVV EDLNSREQETCQPVHVVNVDIQDNHEEATLGAFLICELCQCIQHTEDMENEIDELLQEFE EKSGRTFLHTVCFY |
| 709 |
Q9BQP7 (MGME1) |
 Estimated TM-score=0.925 |
>Mitochondrial genome maintenance exonuclease 1 (Length=344) MKMKLFQTICRQLRSSKFSVESAALVAFSTSSYSCGRKKKVNPYEEVDQEKYSNLVQSVL SSRGVAQTPGSVEEDALLCGPVSKHKLPNQGEDRRVPQNWFPIFNPERSDKPNASDPSVP LKIPLQRNVIPSVTRVLQQTMTKQQVFLLERWKQRMILELGEDGFKEYTSNVFLQGKRFH EALESILSPQETLKERDENLLKSGYIESVQHILKDVSGVRALESAVQHETLNYIGLLDCV AEYQGKLCVIDWKTSEKPKPFIQSTFDNPLQVVAYMGAMNHDTNYSFQVQCGLIVVAYKD GSPAHPHFMDAELCSQYWTKWLLRLEEYTEKKKNQNIQKPEYSE |
| 710 |
O00244 (ATOX1) |
 Estimated TM-score=0.924 |
>Copper transport protein ATOX1 (Length=68) MPKHEFSVDMTCGGCAEAVSRVLNKLGGVKYDIDLPNKKVCIESEHSMDTLLATLKKTGK TVSYLGLE |
| 711 |
Q14353 (GAMT) |
 Estimated TM-score=0.924 |
>Guanidinoacetate N-methyltransferase (Length=236) MSAPSATPIFAPGENCSPAWGAAPAAYDAADTHLRILGKPVMERWETPYMHALAAAASSK GGRVLEVGFGMAIAASKVQEAPIDEHWIIECNDGVFQRLRDWAPRQTHKVIPLKGLWEDV APTLPDGHFDGILYDTYPLSEETWHTHQFNFIKNHAFRLLKPGGVLTYCNLTSWGELMKS KYSDITIMFEETQVPALLEAGFRRENIRTEVMALVPPADCRYYAFPQMITPLVTKG |
| 712 |
Q9GZQ8 (MAP1LC3B) |
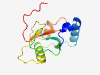 Estimated TM-score=0.924 |
>Microtubule-associated proteins 1A/1B light chain 3B (Length=125) MPSEKTFKQRRTFEQRVEDVRLIREQHPTKIPVIIERYKGEKQLPVLDKTKFLVPDHVNM SELIKIIRRRLQLNANQAFFLLVNGHSMVSVSTPISEVYESEKDEDGFLYMVYASQETFG MKLSV |
| 713 |
Q9UI17 (DMGDH) |
 Estimated TM-score=0.924 |
>Dimethylglycine dehydrogenase, mitochondrial (Length=866) MLRPGAQLLRGLLLRSCPLQGSPGRPRSVCGREGEEKPPLSAETQWKDRAETVIIGGGCV GVSLAYHLAKAGMKDVVLLEKSELTAGSTWHAAGLTTYFHPGINLKKIHYDSIKLYEKLE EETGQVVGFHQPGSIRLATTPVRVDEFKYQMTRTGWHATEQYLIEPEKIQEMFPLLNMNK VLAGLYNPGDGHIDPYSLTMALAAGARKCGALLKYPAPVTSLKARSDGTWDVETPQGSMR ANRIVNAAGFWAREVGKMIGLEHPLIPVQHQYVVTSTISEVKALKRELPVLRDLEGSYYL RQERDGLLFGPYESQEKMKVQDSWVTNGVPPGFGKELFESDLDRIMEHIKAAMEMVPVLK KADIINVVNGPITYSPDILPMVGPHQGVRNYWVAIGFGYGIIHAGGVGKYLSDWILHGEP PFDLIELDPNRYGKWTTTQYTEAKARESYGFNNIVGYPKEERFAGRPTQRVSGLYQRLES KCSMGFHAGWEQPHWFYKPGQDTQYRPSFRRTNWFEPVGSEYKQVMQRVAVTDLSPFGKF NIKGQDSIRLLDHLFANVIPKVGFTNISHMLTPKGRVYAELTVSHQSPGEFLLITGSGSE LHDLRWIEEEAVKGGYDVEIKNITDELGVLGVAGPQARKVLQKLTSEDLSDDVFKFLQTK SLKVSNIPVTAIRISYTGELGWELYHRREDSVALYDAIMNAGQEEGIDNFGTYAMNALRL EKAFRAWGLEMNCDTNPLEAGLEYFVKLNKPADFIGKQALKQIKAKGLKRRLVCLTLATD DVDPEGNESIWYNGKVVGNTTSGSYSYSIQKSLAFAYVPVQLSEVGQQVEVELLGKNYPA VIIQEPLVLTEPTRNRLQKKGGKDKT |
| 714 |
Q8N6N7 (ACBD7) |
 Estimated TM-score=0.924 |
>Acyl-CoA-binding domain-containing protein 7 (Length=88) MALQADFDRAAEDVRKLKARPDDGELKELYGLYKQAIVGDINIACPGMLDLKGKAKWEAW NLKKGLSTEDATSAYISKAKELIEKYGI |
| 715 |
O75936 (BBOX1) |
 Estimated TM-score=0.924 |
>Gamma-butyrobetaine dioxygenase (Length=387) MACTIQKAEALDGAHLMQILWYDEEESLYPAVWLRDNCPCSDCYLDSAKARKLLVEALDV NIGIKGLIFDRKKVYITWPDEHYSEFQADWLKKRCFSKQARAKLQRELFFPECQYWGSEL QLPTLDFEDVLRYDEHAYKWLSTLKKVGIVRLTGASDKPGEVSKLGKRMGFLYLTFYGHT WQVQDKIDANNVAYTTGKLSFHTDYPALHHPPGVQLLHCIKQTVTGGDSEIVDGFNVCQK LKKNNPQAFQILSSTFVDFTDIGVDYCDFSVQSKHKIIELDDKGQVVRINFNNATRDTIF DVPVERVQPFYAALKEFVDLMNSKESKFTFKMNPGDVITFDNWRLLHGRRSYEAGTEISR HLEGAYADWDVVMSRLRILRQRVENGN |
| 716 |
Q9NR31 (SAR1A) |
 Estimated TM-score=0.924 |
>GTP-binding protein SAR1a (Length=198) MSFIFEWIYNGFSSVLQFLGLYKKSGKLVFLGLDNAGKTTLLHMLKDDRLGQHVPTLHPT SEELTIAGMTFTTFDLGGHEQARRVWKNYLPAINGIVFLVDCADHSRLVESKVELNALMT DETISNVPILILGNKIDRTDAISEEKLREIFGLYGQTTGKGNVTLKELNARPMEVFMCSV LKRQGYGEGFRWLSQYID |
| 717 |
P63261 (ACTG1) |
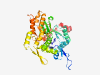 Estimated TM-score=0.924 |
>Actin, cytoplasmic 2 (Length=375) MEEEIAALVIDNGSGMCKAGFAGDDAPRAVFPSIVGRPRHQGVMVGMGQKDSYVGDEAQS KRGILTLKYPIEHGIVTNWDDMEKIWHHTFYNELRVAPEEHPVLLTEAPLNPKANREKMT QIMFETFNTPAMYVAIQAVLSLYASGRTTGIVMDSGDGVTHTVPIYEGYALPHAILRLDL AGRDLTDYLMKILTERGYSFTTTAEREIVRDIKEKLCYVALDFEQEMATAASSSSLEKSY ELPDGQVITIGNERFRCPEALFQPSFLGMESCGIHETTFNSIMKCDVDIRKDLYANTVLS GGTTMYPGIADRMQKEITALAPSTMKIKIIAPPERKYSVWIGGSILASLSTFQQMWISKQ EYDESGPSIVHRKCF |
| 718 |
P10599 (TXN) |
 Estimated TM-score=0.924 |
>Thioredoxin (Length=105) MVKQIESKTAFQEALDAAGDKLVVVDFSATWCGPCKMIKPFFHSLSEKYSNVIFLEVDVD DCQDVASECEVKCMPTFQFFKKGQKVGEFSGANKEKLEATINELV |
| 719 |
P08243 (ASNS) |
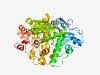 Estimated TM-score=0.924 |
>Asparagine synthetase [glutamine-hydrolyzing] (Length=561) MCGIWALFGSDDCLSVQCLSAMKIAHRGPDAFRFENVNGYTNCCFGFHRLAVVDPLFGMQ PIRVKKYPYLWLCYNGEIYNHKKMQQHFEFEYQTKVDGEIILHLYDKGGIEQTICMLDGV FAFVLLDTANKKVFLGRDTYGVRPLFKAMTEDGFLAVCSEAKGLVTLKHSATPFLKVEPF LPGHYEVLDLKPNGKVASVEMVKYHHCRDVPLHALYDNVEKLFPGFEIETVKNNLRILFN NAVKKRLMTDRRIGCLLSGGLDSSLVAATLLKQLKEAQVQYPLQTFAIGMEDSPDLLAAR KVADHIGSEHYEVLFNSEEGIQALDEVIFSLETYDITTVRASVGMYLISKYIRKNTDSVV IFSGEGSDELTQGYIYFHKAPSPEKAEEESERLLRELYLFDVLRADRTTAAHGLELRVPF LDHRFSSYYLSLPPEMRIPKNGIEKHLLRETFEDSNLIPKEILWRPKEAFSDGITSVKNS WFKILQEYVEHQVDDAMMANAAQKFPFNTPKTKEGYYYRQVFERHYPGRADWLSHYWMPK WINATDPSARTLTHYKSAVKA |
| 720 |
O00399 (DCTN6) |
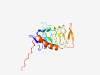 Estimated TM-score=0.924 |
>Dynactin subunit 6 (Length=190) MAEKTQKSVKIAPGAVVCVESEIRGDVTIGPRTVIHPKARIIAEAGPIVIGEGNLIEEQA LIINAYPDNITPDTEDPEPKPMIIGTNNVFEVGCYSQAMKMGDNNVIESKAYVGRNVILT SGCIIGACCNLNTFEVIPENTVIYGADCLRRVQTERPQPQTLQLDFLMKILPNYHHLKKT MKGSSTPVKN |
| 721 |
P36952 (SERPINB5) |
 Estimated TM-score=0.924 |
>Serpin B5 (Length=375) MDALQLANSAFAVDLFKQLCEKEPLGNVLFSPICLSTSLSLAQVGAKGDTANEIGQVLHF ENVKDVPFGFQTVTSDVNKLSSFYSLKLIKRLYVDKSLNLSTEFISSTKRPYAKELETVD FKDKLEETKGQINNSIKDLTDGHFENILADNSVNDQTKILVVNAAYFVGKWMKKFSESET KECPFRVNKTDTKPVQMMNMEATFCMGNIDSINCKIIELPFQNKHLSMFILLPKDVEDES TGLEKIEKQLNSESLSQWTNPSTMANAKVKLSIPKFKVEKMIDPKACLENLGLKHIFSED TSDFSGMSETKGVALSNVIHKVCLEITEDGGDSIEVPGARILQHKDELNADHPFIYIIRH NKTRNIIFFGKFCSP |
| 722 |
O95484 (CLDN9) |
 Estimated TM-score=0.923 |
>Claudin-9 (Length=217) MASTGLELLGMTLAVLGWLGTLVSCALPLWKVTAFIGNSIVVAQVVWEGLWMSCVVQSTG QMQCKVYDSLLALPQDLQAARALCVIALLLALLGLLVAITGAQCTTCVEDEGAKARIVLT AGVILLLAGILVLIPVCWTAHAIIQDFYNPLVAEALKRELGASLYLGWAAAALLMLGGGL LCCTCPPPQVERPRGPRLGYSIPSRSGASGLDKRDYV |
| 723 |
P51580 (TPMT) |
 Estimated TM-score=0.923 |
>Thiopurine S-methyltransferase (Length=245) MDGTRTSLDIEEYSDTEVQKNQVLTLEEWQDKWVNGKTAFHQEQGHQLLKKHLDTFLKGK SGLRVFFPLCGKAVEMKWFADRGHSVVGVEISELGIQEFFTEQNLSYSEEPITEIPGTKV FKSSSGNISLYCCSIFDLPRTNIGKFDMIWDRGALVAINPGDRKCYADTMFSLLGKKFQY LLCVLSYDPTKHPGPPFYVPHAEIERLFGKICNIRCLEKVDAFEERHKSWGIDCLFEKLY LLTEK |
| 724 |
P16050 (ALOX15) |
 Estimated TM-score=0.923 |
>Arachidonate 15-lipoxygenase (Length=662) MGLYRIRVSTGASLYAGSNNQVQLWLVGQHGEAALGKRLWPARGKETELKVEVPEYLGPL LFVKLRKRHLLKDDAWFCNWISVQGPGAGDEVRFPCYRWVEGNGVLSLPEGTGRTVGEDP QGLFQKHREEELEERRKLYRWGNWKDGLILNMAGAKLYDLPVDERFLEDKRVDFEVSLAK GLADLAIKDSLNVLTCWKDLDDFNRIFWCGQSKLAERVRDSWKEDALFGYQFLNGANPVV LRRSAHLPARLVFPPGMEELQAQLEKELEGGTLFEADFSLLDGIKANVILCSQQHLAAPL VMLKLQPDGKLLPMVIQLQLPRTGSPPPPLFLPTDPPMAWLLAKCWVRSSDFQLHELQSH LLRGHLMAEVIVVATMRCLPSIHPIFKLIIPHLRYTLEINVRARTGLVSDMGIFDQIMST GGGGHVQLLKQAGAFLTYSSFCPPDDLADRGLLGVKSSFYAQDALRLWEIIYRYVEGIVS LHYKTDVAVKDDPELQTWCREITEIGLQGAQDRGFPVSLQARDQVCHFVTMCIFTCTGQH ASVHLGQLDWYSWVPNAPCTMRLPPPTTKDATLETVMATLPNFHQASLQMSITWQLGRRQ PVMVAVGQHEEEYFSGPEPKAVLKKFREELAALDKEIEIRNAKLDMPYEYLRPSVVENSV AI |
| 725 |
Q15391 (P2RY14) |
 Estimated TM-score=0.923 |
>P2Y purinoceptor 14 (Length=338) MINSTSTQPPDESCSQNLLITQQIIPVLYCMVFIAGILLNGVSGWIFFYVPSSKSFIIYL KNIVIADFVMSLTFPFKILGDSGLGPWQLNVFVCRVSAVLFYVNMYVSIVFFGLISFDRY YKIVKPLWTSFIQSVSYSKLLSVIVWMLMLLLAVPNIILTNQSVREVTQIKCIELKSELG RKWHKASNYIFVAIFWIVFLLLIVFYTAITKKIFKSHLKSSRNSTSVKKKSSRNIFSIVF VFFVCFVPYHIARIPYTKSQTEAHYSCQSKEILRYMKEFTLLLSAANVCLDPIIYFFLCQ PFREILCKKLHIPLKAQNDLDISRIKRGNTTLESTDTL |
| 726 |
Q9H7X0 (NAA60) |
 Estimated TM-score=0.923 |
>N-alpha-acetyltransferase 60 (Length=242) MTEVVPSSALSEVSLRLLCHDDIDTVKHLCGDWFPIEYPDSWYRDITSNKKFFSLAATYR GAIVGMIVAEIKNRTKIHKEDGDILASNFSVDTQVAYILSLGVVKEFRKHGIGSLLLESL KDHISTTAQDHCKAIYLHVLTTNNTAINFYENRDFKQHHYLPYYYSIRGVLKDGFTYVLY INGGHPPWTILDYIQHLGSALASLSPCSIPHRVYRQAHSLLCSFLPWSGISSKSGIEYSR TM |
| 727 |
Q9BZX2 (UCK2) |
 Estimated TM-score=0.923 |
>Uridine-cytidine kinase 2 (Length=261) MAGDSEQTLQNHQQPNGGEPFLIGVSGGTASGKSSVCAKIVQLLGQNEVDYRQKQVVILS QDSFYRVLTSEQKAKALKGQFNFDHPDAFDNELILKTLKEITEGKTVQIPVYDFVSHSRK EETVTVYPADVVLFEGILAFYSQEVRDLFQMKLFVDTDADTRLSRRVLRDISERGRDLEQ ILSQYITFVKPAFEEFCLPTKKYADVIIPRGADNLVAINLIVQHIQDILNGGPSKRQTNG CLNGYTPSRKRQASESSSRPH |
| 728 |
O95372 (LYPLA2) |
 Estimated TM-score=0.922 |
>Acyl-protein thioesterase 2 (Length=231) MCGNTMSVPLLTDAATVSGAERETAAVIFLHGLGDTGHSWADALSTIRLPHVKYICPHAP RIPVTLNMKMVMPSWFDLMGLSPDAPEDEAGIKKAAENIKALIEHEMKNGIPANRIVLGG FSQGGALSLYTALTCPHPLAGIVALSCWLPLHRAFPQAANGSAKDLAILQCHGELDPMVP VRFGALTAEKLRSVVTPARVQFKTYPGVMHSSCPQEMAAVKEFLEKLLPPV |
| 729 |
P84098 (RPL19) |
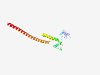 Estimated TM-score=0.922 |
>60S ribosomal protein L19 (Length=196) MSMLRLQKRLASSVLRCGKKKVWLDPNETNEIANANSRQQIRKLIKDGLIIRKPVTVHSR ARCRKNTLARRKGRHMGIGKRKGTANARMPEKVTWMRRMRILRRLLRRYRESKKIDRHMY HSLYLKVKGNVFKNKRILMEHIHKLKADKARKKLLADQAEARRSKTKEARKRREERLQAK KEEIIKTLSKEEETKK |
| 730 |
O15296 (ALOX15B) |
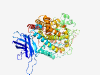 Estimated TM-score=0.922 |
>Arachidonate 15-lipoxygenase B (Length=676) MAEFRVRVSTGEAFGAGTWDKVSVSIVGTRGESPPLPLDNLGKEFTAGAEEDFQVTLPED VGRVLLLRVHKAPPVLPLLGPLAPDAWFCRWFQLTPPRGGHLLFPCYQWLEGAGTLVLQE GTAKVSWADHHPVLQQQRQEELQARQEMYQWKAYNPGWPHCLDEKTVEDLELNIKYSTAK NANFYLQAGSAFAEMKIKGLLDRKGLWRSLNEMKRIFNFRRTPAAEHAFEHWQEDAFFAS QFLNGLNPVLIRRCHYLPKNFPVTDAMVASVLGPGTSLQAELEKGSLFLVDHGILSGIQT NVINGKPQFSAAPMTLLYQSPGCGPLLPLAIQLSQTPGPNSPIFLPTDDKWDWLLAKTWV RNAEFSFHEALTHLLHSHLLPEVFTLATLRQLPHCHPLFKLLIPHTRYTLHINTLARELL IVPGQVVDRSTGIGIEGFSELIQRNMKQLNYSLLCLPEDIRTRGVEDIPGYYYRDDGMQI WGAVERFVSEIIGIYYPSDESVQDDRELQAWVREIFSKGFLNQESSGIPSSLETREALVQ YVTMVIFTCSAKHAAVSAGQFDSCAWMPNLPPSMQLPPPTSKGLATCEGFIATLPPVNAT CDVILALWLLSKEPGDQRPLGTYPDEHFTEEAPRRSIATFQSRLAQISRGIQERNQGLVL PYTYLDPPLIENSVSI |
| 731 |
P51398 (DAP3) |
 Estimated TM-score=0.922 |
>28S ribosomal protein S29, mitochondrial (Length=398) MMLKGITRLISRIHKLDPGRFLHMGTQARQSIAAHLDNQVPVESPRAISRTNENDPAKHG DQHEGQHYNISPQDLETVFPHGLPPRFVMQVKTFSEACLMVRKPALELLHYLKNTSFAYP AIRYLLYGEKGTGKTLSLCHVIHFCAKQDWLILHIPDAHLWVKNCRDLLQSSYNKQRFDQ PLEASTWLKNFKTTNERFLNQIKVQEKYVWNKRESTEKGSPLGEVVEQGITRVRNATDAV GIVLKELKRQSSLGMFHLLVAVDGINALWGRTTLKREDKSPIAPEELALVHNLRKMMKND WHGGAIVSALSQTGSLFKPRKAYLPQELLGKEGFDALDPFIPILVSNYNPKEFESCIQYY LENNWLQHEKAPTEEGKKELLFLSNANPSLLERHCAYL |
| 732 |
Q9BRX5 (GINS3) |
 Estimated TM-score=0.922 |
>DNA replication complex GINS protein PSF3 (Length=216) MSEAYFRVESGALGPEENFLSLDDILMSHEKLPVRTETAMPRLGAFFLERSAGAETDNAV PQGSKLELPLWLAKGLFDNKRRILSVELPKIYQEGWRTVFSADPNVVDLHKMGPHFYGFG SQLLHFDSPENADISQSLLQTFIGRFRRIMDSSQNAYNEDTSALVARLDEMERGLFQTGQ KGLNDFQCWEKGQASQITASNLVQNYKKRKFTDMED |
| 733 |
Q96N28 (PRELID3A) |
 Estimated TM-score=0.922 |
>PRELI domain containing protein 3A (Length=172) MKIWSSEHVFGHPWDTVIQAAMRKYPNPMNPSVLGVDVLQRRVDGRGRLHSLRLLSTEWG LPSLVRAILGTSRTLTYIREHSVVDPVEKKMELCSTNITLTNLVSVNERLVYTPHPENPE MTVLTQEAIITVKGISLGSYLESLMANTISSNAKKGWAAIEWIIEHSESAVS |
| 734 |
P11413 (G6PD) |
 Estimated TM-score=0.922 |
>Glucose-6-phosphate 1-dehydrogenase (Length=515) MAEQVALSRTQVCGILREELFQGDAFHQSDTHIFIIMGASGDLAKKKIYPTIWWLFRDGL LPENTFIVGYARSRLTVADIRKQSEPFFKATPEEKLKLEDFFARNSYVAGQYDDAASYQR LNSHMNALHLGSQANRLFYLALPPTVYEAVTKNIHESCMSQIGWNRIIVEKPFGRDLQSS DRLSNHISSLFREDQIYRIDHYLGKEMVQNLMVLRFANRIFGPIWNRDNIACVILTFKEP FGTEGRGGYFDEFGIIRDVMQNHLLQMLCLVAMEKPASTNSDDVRDEKVKVLKCISEVQA NNVVLGQYVGNPDGEGEATKGYLDDPTVPRGSTTATFAAVVLYVENERWDGVPFILRCGK ALNERKAEVRLQFHDVAGDIFHQQCKRNELVIRVQPNEAVYTKMMTKKPGMFFNPEESEL DLTYGNRYKNVKLPDAYERLILDVFCGSQMHFVRSDELREAWRIFTPLLHQIELEKPKPI PYIYGSRGPTEADELMKRVGFQYEGTYKWVNPHKL |
| 735 |
P49356 (FNTB) |
 Estimated TM-score=0.922 |
>Protein farnesyltransferase subunit beta (Length=437) MASPSSFTYYCPPSSSPVWSEPLYSLRPEHARERLQDDSVETVTSIEQAKVEEKIQEVFS SYKFNHLVPRLVLQREKHFHYLKRGLRQLTDAYECLDASRPWLCYWILHSLELLDEPIPQ IVATDVCQFLELCQSPEGGFGGGPGQYPHLAPTYAAVNALCIIGTEEAYDIINREKLLQY LYSLKQPDGSFLMHVGGEVDVRSAYCAASVASLTNIITPDLFEGTAEWIARCQNWEGGIG GVPGMEAHGGYTFCGLAALVILKRERSLNLKSLLQWVTSRQMRFEGGFQGRCNKLVDGCY SFWQAGLLPLLHRALHAQGDPALSMSHWMFHQQALQEYILMCCQCPAGGLLDKPGKSRDF YHTCYCLSGLSIAQHFGSGAMLHDVVLGVPENALQPTHPVYNIGPDKVIQATTYFLQKPV PGFEELKDETSAEPATD |
| 736 |
P47895 (ALDH1A3) |
 Estimated TM-score=0.922 |
>Aldehyde dehydrogenase family 1 member A3 (Length=512) MATANGAVENGQPDRKPPALPRPIRNLEVKFTKIFINNEWHESKSGKKFATCNPSTREQI CEVEEGDKPDVDKAVEAAQVAFQRGSPWRRLDALSRGRLLHQLADLVERDRATLAALETM DTGKPFLHAFFIDLEGCIRTLRYFAGWADKIQGKTIPTDDNVVCFTRHEPIGVCGAITPW NFPLLMLVWKLAPALCCGNTMVLKPAEQTPLTALYLGSLIKEAGFPPGVVNIVPGFGPTV GAAISSHPQINKIAFTGSTEVGKLVKEAASRSNLKRVTLELGGKNPCIVCADADLDLAVE CAHQGVFFNQGQCCTAASRVFVEEQVYSEFVRRSVEYAKKRPVGDPFDVKTEQGPQIDQK QFDKILELIESGKKEGAKLECGGSAMEDKGLFIKPTVFSEVTDNMRIAKEEIFGPVQPIL KFKSIEEVIKRANSTDYGLTAAVFTKNLDKALKLASALESGTVWINCYNALYAQAPFGGF KMSGNGRELGEYALAEYTEVKTVTIKLGDKNP |
| 737 |
P05413 (FABP3) |
 Estimated TM-score=0.921 |
>Fatty acid-binding protein, heart (Length=133) MVDAFLGTWKLVDSKNFDDYMKSLGVGFATRQVASMTKPTTIIEKNGDILTLKTHSTFKN TEISFKLGVEFDETTADDRKVKSIVTLDGGKLVHLQKWDGQETTLVRELIDGKLILTLTH GTAVCTRTYEKEA |
| 738 |
Q9NRN9 (METTL5) |
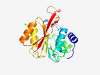 Estimated TM-score=0.921 |
>rRNA N6-adenosine-methyltransferase METTL5 (Length=209) MKKVRLKELESRLQQVDGFEKPKLLLEQYPTRPHIAACMLYTIHNTYDDIENKVVADLGC GCGVLSIGTAMLGAGLCVGFDIDEDALEIFNRNAEEFELTNIDMVQCDVCLLSNRMSKSF DTVIMNPPFGTKNNKGTDMAFLKTALEMARTAVYSLHKSSTREHVQKKAAEWKIKIDIIA ELRYDLPASYKFHKKKSVDIEVDLIRFSF |
| 739 |
P04001 (OPN1MW) |
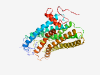 Estimated TM-score=0.921 |
>Medium-wave-sensitive opsin 1 (Length=364) MAQQWSLQRLAGRHPQDSYEDSTQSSIFTYTNSNSTRGPFEGPNYHIAPRWVYHLTSVWM IFVVIASVFTNGLVLAATMKFKKLRHPLNWILVNLAVADLAETVIASTISVVNQVYGYFV LGHPMCVLEGYTVSLCGITGLWSLAIISWERWMVVCKPFGNVRFDAKLAIVGIAFSWIWA AVWTAPPIFGWSRYWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMVTCCITPLSIIVLCYL QVWLAIRAVAKQQKESESTQKAEKEVTRMVVVMVLAFCFCWGPYAFFACFAAANPGYPFH PLMAALPAFFAKSATIYNPVIYVFMNRQFRNCILQLFGKKVDDGSELSSASKTEVSSVSS VSPA |
| 740 |
Q16678 (CYP1B1) |
 Estimated TM-score=0.921 |
>Cytochrome P450 1B1 (Length=543) MGTSLSPNDPWPLNPLSIQQTTLLLLLSVLATVHVGQRLLRQRRRQLRSAPPGPFAWPLI GNAAAVGQAAHLSFARLARRYGDVFQIRLGSCPIVVLNGERAIHQALVQQGSAFADRPAF ASFRVVSGGRSMAFGHYSEHWKVQRRAAHSMMRNFFTRQPRSRQVLEGHVLSEARELVAL LVRGSADGAFLDPRPLTVVAVANVMSAVCFGCRYSHDDPEFRELLSHNEEFGRTVGAGSL VDVMPWLQYFPNPVRTVFREFEQLNRNFSNFILDKFLRHCESLRPGAAPRDMMDAFILSA EKKAAGDSHGGGARLDLENVPATITDIFGASQDTLSTALQWLLLLFTRYPDVQTRVQAEL DQVVGRDRLPCMGDQPNLPYVLAFLYEAMRFSSFVPVTIPHATTANTSVLGYHIPKDTVV FVNQWSVNHDPLKWPNPENFDPARFLDKDGLINKDLTSRVMIFSVGKRRCIGEELSKMQL FLFISILAHQCDFRANPNEPAKMNFSYGLTIKPKSFKVNVTLRESMELLDSAVQNLQAKE TCQ |
| 741 |
P27216 (ANXA13) |
 Estimated TM-score=0.921 |
>Annexin A13 (Length=316) MGNRHAKASSPQGFDVDRDAKKLNKACKGMGTNEAAIIEILSGRTSDERQQIKQKYKATY GKELEEVLKSELSGNFEKTALALLDRPSEYAARQLQKAMKGLGTDESVLIEVLCTRTNKE IIAIKEAYQRLFDRSLESDVKGDTSGNLKKILVSLLQANRNEGDDVDKDLAGQDAKDLYD AGEGRWGTDELAFNEVLAKRSYKQLRATFQAYQILIGKDIEEAIEEETSGDLQKAYLTLV RCAQDCEDYFAERLYKSMKGAGTDEETLIRIVVTRAEVDLQGIKAKFQEKYQKSLSDMVR SDTSGDFRKLLVALLH |
| 742 |
Q93096 (PTP4A1) |
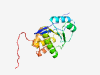 Estimated TM-score=0.921 |
>Protein tyrosine phosphatase type IVA 1 (Length=173) MARMNRPAPVEVTYKNMRFLITHNPTNATLNKFIEELKKYGVTTIVRVCEATYDTTLVEK EGIHVLDWPFDDGAPPSNQIVDDWLSLVKIKFREEPGCCIAVHCVAGLGRAPVLVALALI EGGMKYEDAVQFIRQKRRGAFNSKQLLYLEKYRPKMRLRFKDSNGHRNNCCIQ |
| 743 |
Q96E14 (RMI2) |
 Estimated TM-score=0.921 |
>RecQ-mediated genome instability protein 2 (Length=147) MAAAADSFSGGPAGVRLPRSPPLKVLAEQLRRDAEGGPGAWRLSRAAAGRGPLDLAAVWM QGRVVMADRGEARLRDPSGDFSVRGLERVPRGRPCLVPGKYVMVMGVVQACSPEPCLQAV KMTDLSDNPIHESMWELEVEDLHRNIP |
| 744 |
P62487 (POLR2G) |
 Estimated TM-score=0.921 |
>DNA-directed RNA polymerase II subunit RPB7 (Length=172) MFYHISLEHEILLHPRYFGPNLLNTVKQKLFTEVEGTCTGKYGFVIAVTTIDNIGAGVIQ PGRGFVLYPVKYKAIVFRPFKGEVVDAVVTQVNKVGLFTEIGPMSCFISRHSIPSEMEFD PNSNPPCYKTMDEDIVIQQDDEIRLKIVGTRVDKNDIFAIGSLMDDYLGLVS |
| 745 |
O00299 (CLIC1) |
 Estimated TM-score=0.921 |
>Chloride intracellular channel protein 1 (Length=241) MAEEQPQVELFVKAGSDGAKIGNCPFSQRLFMVLWLKGVTFNVTTVDTKRRTETVQKLCP GGQLPFLLYGTEVHTDTNKIEEFLEAVLCPPRYPKLAALNPESNTAGLDIFAKFSAYIKN SNPALNDNLEKGLLKALKVLDNYLTSPLPEEVDETSAEDEGVSQRKFLDGNELTLADCNL LPKLHIVQVVCKKYRGFTIPEAFRGVHRYLSNAYAREEFASTCPDDEEIELAYEQVAKAL K |
| 746 |
Q14392 (LRRC32) |
 Estimated TM-score=0.921 |
>Transforming growth factor beta activator LRRC32 (Length=662) MRPQILLLLALLTLGLAAQHQDKVPCKMVDKKVSCQVLGLLQVPSVLPPDTETLDLSGNQ LRSILASPLGFYTALRHLDLSTNEISFLQPGAFQALTHLEHLSLAHNRLAMATALSAGGL GPLPRVTSLDLSGNSLYSGLLERLLGEAPSLHTLSLAENSLTRLTRHTFRDMPALEQLDL HSNVLMDIEDGAFEGLPRLTHLNLSRNSLTCISDFSLQQLRVLDLSCNSIEAFQTASQPQ AEFQLTWLDLRENKLLHFPDLAALPRLIYLNLSNNLIRLPTGPPQDSKGIHAPSEGWSAL PLSAPSGNASGRPLSQLLNLDLSYNEIELIPDSFLEHLTSLCFLNLSRNCLRTFEARRLG SLPCLMLLDLSHNALETLELGARALGSLRTLLLQGNALRDLPPYTFANLASLQRLNLQGN RVSPCGGPDEPGPSGCVAFSGITSLRSLSLVDNEIELLRAGAFLHTPLTELDLSSNPGLE VATGALGGLEASLEVLALQGNGLMVLQVDLPCFICLKRLNLAENRLSHLPAWTQAVSLEV LDLRNNSFSLLPGSAMGGLETSLRRLYLQGNPLSCCGNGWLAAQLHQGRVDVDATQDLIC RFSSQEEVSLSHVRPEDCEKGGLKNINLIIILTFILVSAILLTTLAACCCVRRQKFNQQY KA |
| 747 |
Q9NP97 (DYNLRB1) |
 Estimated TM-score=0.920 |
>Dynein light chain roadblock-type 1 (Length=96) MAEVEETLKRLQSQKGVQGIIVVNTEGIPIKSTMDNPTTTQYASLMHSFILKARSTVRDI DPQNDLTFLRIRSKKNEIMVAPDKDYFLIVIQNPTE |
| 748 |
P43487 (RANBP1) |
 Estimated TM-score=0.920 |
>Ran-specific GTPase-activating protein (Length=201) MAAAKDTHEDHDTSTENTDESNHDPQFEPIVSLPEQEIKTLEEDEEELFKMRAKLFRFAS ENDLPEWKERGTGDVKLLKHKEKGAIRLLMRRDKTLKICANHYITPMMELKPNAGSDRAW VWNTHADFADECPKPELLAIRFLNAENAQKFKTKFEECRKEIEEREKKAGSGKNDHAEKV AEKLEALSVKEETKEDAEEKQ |
| 749 |
Q15126 (PMVK) |
 Estimated TM-score=0.920 |
>Phosphomevalonate kinase (Length=192) MAPLGGAPRLVLLFSGKRKSGKDFVTEALQSRLGADVCAVLRLSGPLKEQYAQEHGLNFQ RLLDTSTYKEAFRKDMIRWGEEKRQADPGFFCRKIVEGISQPIWLVSDTRRVSDIQWFRE AYGAVTQTVRVVALEQSRQQRGWVFTPGVDDAESECGLDNFGDFDWVIENHGVEQRLEEQ LENLIEFIRSRL |
| 750 |
P10746 (UROS) |
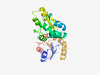 Estimated TM-score=0.920 |
>Uroporphyrinogen-III synthase (Length=265) MKVLLLKDAKEDDCGQDPYIRELGLYGLEATLIPVLSFEFLSLPSFSEKLSHPEDYGGLI FTSPRAVEAAELCLEQNNKTEVWERSLKEKWNAKSVYVVGNATASLVSKIGLDTEGETCG NAEKLAEYICSRESSALPLLFPCGNLKREILPKALKDKGIAMESITVYQTVAHPGIQGNL NSYYSQQGVPASITFFSPSGLTYSLKHIQELSGDNIDQIKFAAIGPTTARALAAQGLPVS CTAESPTPQALATGIRKALQPHGCC |
| 751 |
P62277 (RPS13) |
 Estimated TM-score=0.920 |
>40S ribosomal protein S13 (Length=151) MGRMHAPGKGLSQSALPYRRSVPTWLKLTSDDVKEQIYKLAKKGLTPSQIGVILRDSHGV AQVRFVTGNKILRILKSKGLAPDLPEDLYHLIKKAVAVRKHLERNRKDKDAKFRLILIES RIHRLARYYKTKRVLPPNWKYESSTASALVA |
| 752 |
P49419 (ALDH7A1) |
 Estimated TM-score=0.920 |
>Alpha-aminoadipic semialdehyde dehydrogenase (Length=539) MWRLPRALCVHAAKTSKLSGPWSRPAAFMSTLLINQPQYAWLKELGLREENEGVYNGSWG GRGEVITTYCPANNEPIARVRQASVADYEETVKKAREAWKIWADIPAPKRGEIVRQIGDA LREKIQVLGSLVSLEMGKILVEGVGEVQEYVDICDYAVGLSRMIGGPILPSERSGHALIE QWNPVGLVGIITAFNFPVAVYGWNNAIAMICGNVCLWKGAPTTSLISVAVTKIIAKVLED NKLPGAICSLTCGGADIGTAMAKDERVNLLSFTGSTQVGKQVGLMVQERFGRSLLELGGN NAIIAFEDADLSLVVPSALFAAVGTAGQRCTTARRLFIHESIHDEVVNRLKKAYAQIRVG NPWDPNVLYGPLHTKQAVSMFLGAVEEAKKEGGTVVYGGKVMDRPGNYVEPTIVTGLGHD ASIAHTETFAPILYVFKFKNEEEVFAWNNEVKQGLSSSIFTKDLGRIFRWLGPKGSDCGI VNVNIPTSGAEIGGAFGGEKHTGGGRESGSDAWKQYMRRSTCTINYSKDLPLAQGIKFQ |
| 753 |
Q969H6 (POP5) |
 Estimated TM-score=0.920 |
>Ribonuclease P/MRP protein subunit POP5 (Length=163) MVRFKHRYLLCELVSDDPRCRLSLDDRVLSSLVRDTIARVHGTFGAAACSIGFAVRYLNA YTGIVLLRCRKEFYQLVWSALPFITYLENKGHRYPCFFNTLHVGGTIRTCQKFLIQYNRR QLLILLQNCTDEGEREAIQKSVTRSCLLEEEEESGEEAAEAME |
| 754 |
O76054 (SEC14L2) |
 Estimated TM-score=0.920 |
>SEC14-like protein 2 (Length=403) MSGRVGDLSPRQKEALAKFRENVQDVLPALPNPDDYFLLRWLRARSFDLQKSEAMLRKHV EFRKQKDIDNIISWQPPEVIQQYLSGGMCGYDLDGCPVWYDIIGPLDAKGLLFSASKQDL LRTKMRECELLLQECAHQTTKLGRKVETITIIYDCEGLGLKHLWKPAVEAYGEFLCMFEE NYPETLKRLFVVKAPKLFPVAYNLIKPFLSEDTRKKIMVLGANWKEVLLKHISPDQVPVE YGGTMTDPDGNPKCKSKINYGGDIPRKYYVRDQVKQQYEHSVQISRGSSHQVEYEILFPG CVLRWQFMSDGADVGFGIFLKTKMGERQRAGEMTEVLPNQRYNSHLVPEDGTLTCSDPGI YVLRFDNTYSFIHAKKVNFTVEVLLPDKASEEKMKQLGAGTPK |
| 755 |
Q9UM13 (ANAPC10) |
 Estimated TM-score=0.920 |
>Anaphase-promoting complex subunit 10 (Length=185) MTTPNKTPPGADPKQLERTGTVREIGSQAVWSLSSCKPGFGVDQLRDDNLETYWQSDGSQ PHLVNIQFRRKTTVKTLCIYADYKSDESYTPSKISVRVGNNFHNLQEIRQLELVEPSGWI HVPLTDNHKKPTRTFMIQIAVLANHQNGRDTHMRQIKIYTPVEESSIGKFPRCTTIDFMM YRSIR |
| 756 |
P55769 (SNU13) |
 Estimated TM-score=0.920 |
>NHP2-like protein 1 (Length=128) MTEADVNPKAYPLADAHLTKKLLDLVQQSCNYKQLRKGANEATKTLNRGISEFIVMAADA EPLEIILHLPLLCEDKNVPYVFVRSKQALGRACGVSRPVIACSVTIKEGSQLKQQIQSIQ QSIERLLV |
| 757 |
Q86TI2 (DPP9) |
 Estimated TM-score=0.919 |
>Dipeptidyl peptidase 9 (Length=863) MATTGTPTADRGDAAATDDPAARFQVQKHSWDGLRSIIHGSRKYSGLIVNKAPHDFQFVQ KTDESGPHSHRLYYLGMPYGSRENSLLYSEIPKKVRKEALLLLSWKQMLDHFQATPHHGV YSREEELLRERKRLGVFGITSYDFHSESGLFLFQASNSLFHCRDGGKNGFMVSPMKPLEI KTQCSGPRMDPKICPADPAFFSFINNSDLWVANIETGEERRLTFCHQGLSNVLDDPKSAG VATFVIQEEFDRFTGYWWCPTASWEGSEGLKTLRILYEEVDESEVEVIHVPSPALEERKT DSYRYPRTGSKNPKIALKLAEFQTDSQGKIVSTQEKELVQPFSSLFPKVEYIARAGWTRD GKYAWAMFLDRPQQWLQLVLLPPALFIPSTENEEQRLASARAVPRNVQPYVVYEEVTNVW INVHDIFYPFPQSEGEDELCFLRANECKTGFCHLYKVTAVLKSQGYDWSEPFSPGEDEFK CPIKEEIALTSGEWEVLARHGSKIWVNEETKLVYFQGTKDTPLEHHLYVVSYEAAGEIVR LTTPGFSHSCSMSQNFDMFVSHYSSVSTPPCVHVYKLSGPDDDPLHKQPRFWASMMEAAS CPPDYVPPEIFHFHTRSDVRLYGMIYKPHALQPGKKHPTVLFVYGGPQVQLVNNSFKGIK YLRLNTLASLGYAVVVIDGRGSCQRGLRFEGALKNQMGQVEIEDQVEGLQFVAEKYGFID LSRVAIHGWSYGGFLSLMGLIHKPQVFKVAIAGAPVTVWMAYDTGYTERYMDVPENNQHG YEAGSVALHVEKLPNEPNRLLILHGFLDENVHFFHTNFLVSQLIRAGKPYQLQIYPNERH SIRCPESGEHYEVTLLHFLQEYL |
| 758 |
P48163 (ME1) |
 Estimated TM-score=0.919 |
>NADP-dependent malic enzyme (Length=572) MEPEAPRRRHTHQRGYLLTRNPHLNKDLAFTLEERQQLNIHGLLPPSFNSQEIQVLRVVK NFEHLNSDFDRYLLLMDLQDRNEKLFYRVLTSDIEKFMPIVYTPTVGLACQQYSLVFRKP RGLFITIHDRGHIASVLNAWPEDVIKAIVVTDGERILGLGDLGCNGMGIPVGKLALYTAC GGMNPQECLPVILDVGTENEELLKDPLYIGLRQRRVRGSEYDDFLDEFMEAVSSKYGMNC LIQFEDFANVNAFRLLNKYRNQYCTFNDDIQGTASVAVAGLLAALRITKNKLSDQTILFQ GAGEAALGIAHLIVMALEKEGLPKEKAIKKIWLVDSKGLIVKGRASLTQEKEKFAHEHEE MKNLEAIVQEIKPTALIGVAAIGGAFSEQILKDMAAFNERPIIFALSNPTSKAECSAEQC YKITKGRAIFASGSPFDPVTLPNGQTLYPGQGNNSYVFPGVALGVVACGLRQITDNIFLT TAEVIAQQVSDKHLEEGRLYPPLNTIRDVSLKIAEKIVKDAYQEKTATVYPEPQNKEAFV RSQMYSTDYDQILPDCYSWPEEVQKIQTKVDQ |
| 759 |
O43709 (BUD23) |
 Estimated TM-score=0.919 |
>Probable 18S rRNA (guanine-N(7))-methyltransferase (Length=281) MASRGRRPEHGGPPELFYDETEARKYVRNSRMIDIQTRMAGRALELLYLPENKPCYLLDI GCGTGLSGSYLSDEGHYWVGLDISPAMLDEAVDREIEGDLLLGDMGQGIPFKPGTFDGCI SISAVQWLCNANKKSENPAKRLYCFFASLFSVLVRGSRAVLQLYPENSEQLELITTQATK AGFSGGMVVDYPNSAKAKKFYLCLFSGPSTFIPEGLSENQDEVEPRESVFTNERFPLRMS RRGMVRKSRAWVLEKKERHRRQGREVRPDTQYTGRKRKPRF |
| 760 |
P04062 (GBA) |
 Estimated TM-score=0.919 |
>Lysosomal acid glucosylceramidase (Length=536) MEFSSPSREECPKPLSRVSIMAGSLTGLLLLQAVSWASGARPCIPKSFGYSSVVCVCNAT YCDSFDPPTFPALGTFSRYESTRSGRRMELSMGPIQANHTGTGLLLTLQPEQKFQKVKGF GGAMTDAAALNILALSPPAQNLLLKSYFSEEGIGYNIIRVPMASCDFSIRTYTYADTPDD FQLHNFSLPEEDTKLKIPLIHRALQLAQRPVSLLASPWTSPTWLKTNGAVNGKGSLKGQP GDIYHQTWARYFVKFLDAYAEHKLQFWAVTAENEPSAGLLSGYPFQCLGFTPEHQRDFIA RDLGPTLANSTHHNVRLLMLDDQRLLLPHWAKVVLTDPEAAKYVHGIAVHWYLDFLAPAK ATLGETHRLFPNTMLFASEACVGSKFWEQSVRLGSWDRGMQYSHSIITNLLYHVVGWTDW NLALNPEGGPNWVRNFVDSPIIVDITKDTFYKQPMFYHLGHFSKFIPEGSQRVGLVASQK NDLDAVALMHPDGSAVVVVLNRSSKDVPLTIKDPAVGFLETISPGYSIHTYLWRRQ |
| 761 |
Q9Y399 (MRPS2) |
 Estimated TM-score=0.919 |
>28S ribosomal protein S2, mitochondrial (Length=296) MATSSAALPRILGAGARAPSRWLGFLGKATPRPARPSRRTLGSATALMIRESEDSTDFND KILNEPLKHSDFFNVKELFSVRSLFDARVHLGHKAGCRHRFMEPYIFGSRLDHDIIDLEQ TATHLQLALNFTAHMAYRKGIILFISRNRQFSYLIENMARDCGEYAHTRYFRGGMLTNAR LLFGPTVRLPDLIIFLHTLNNIFEPHVAVRDAAKMNIPTVGIVDTNCNPCLITYPVPGND DSPLAVHLYCRLFQTAITRAKEKRQQVEALYRLQGQKEPGDQGPAHPPGADMSHSL |
| 762 |
P82664 (MRPS10) |
 Estimated TM-score=0.919 |
>28S ribosomal protein S10, mitochondrial (Length=201) MAARTAFGAVCRRLWQGLGNFSVNTSKGNTAKNGGLLLSTNMKWVQFSNLHVDVPKDLTK PVVTISDEPDILYKRLSVLVKGHDKAVLDSYEYFAVLAAKELGISIKVHEPPRKIERFTL LQSVHIYKKHRVQYEMRTLYRCLELEHLTGSTADVYLEYIQRNLPEGVAMEVTKTQLEQL PEHIKEPIWETLSEEKEESKS |
| 763 |
Q9HD34 (LYRM4) |
 Estimated TM-score=0.919 |
>LYR motif-containing protein 4 (Length=91) MAASSRAQVLSLYRAMLRESKRFSAYNYRTYAVRRIRDAFRENKNVKDPVEIQTLVNKAK RDLGVIRRQVHIGQLYSTDKLIIENRDMPRT |
| 764 |
P02768 (ALB) |
 Estimated TM-score=0.918 |
>Serum albumin (Length=609) MKWVTFISLLFLFSSAYSRGVFRRDAHKSEVAHRFKDLGEENFKALVLIAFAQYLQQCPF EDHVKLVNEVTEFAKTCVADESAENCDKSLHTLFGDKLCTVATLRETYGEMADCCAKQEP ERNECFLQHKDDNPNLPRLVRPEVDVMCTAFHDNEETFLKKYLYEIARRHPYFYAPELLF FAKRYKAAFTECCQAADKAACLLPKLDELRDEGKASSAKQRLKCASLQKFGERAFKAWAV ARLSQRFPKAEFAEVSKLVTDLTKVHTECCHGDLLECADDRADLAKYICENQDSISSKLK ECCEKPLLEKSHCIAEVENDEMPADLPSLAADFVESKDVCKNYAEAKDVFLGMFLYEYAR RHPDYSVVLLLRLAKTYETTLEKCCAAADPHECYAKVFDEFKPLVEEPQNLIKQNCELFE QLGEYKFQNALLVRYTKKVPQVSTPTLVEVSRNLGKVGSKCCKHPEAKRMPCAEDYLSVV LNQLCVLHEKTPVSDRVTKCCTESLVNRRPCFSALEVDETYVPKEFNAETFTFHADICTL SEKERQIKKQTALVELVKHKPKATKEQLKAVMDDFAAFVEKCCKADDKETCFAEEGKKLV AASQAALGL |
| 765 |
P46952 (HAAO) |
 Estimated TM-score=0.918 |
>3-hydroxyanthranilate 3,4-dioxygenase (Length=286) MERRLGVRAWVKENRGSFQPPVCNKLMHQEQLKVMFIGGPNTRKDYHIEEGEEVFYQLEG DMVLRVLEQGKHRDVVIRQGEIFLLPARVPHSPQRFANTVGLVVERRRLETELDGLRYYV GDTMDVLFEKWFYCKDLGTQLAPIIQEFFSSEQYRTGKPIPDQLLKEPPFPLSTRSIMEP MSLDAWLDSHHRELQAGTPLSLFGDTYETQVIAYGQGSSEGLRQNVDVWLWQLEGSSVVT MGGRRLSLAPDDSLLVLAGTSYAWERTQGSVALSVTQDPACKKPLG |
| 766 |
P22090 (RPS4Y1) |
 Estimated TM-score=0.918 |
>40S ribosomal protein S4, Y isoform 1 (Length=263) MARGPKKHLKRVAAPKHWMLDKLTGVFAPRPSTGPHKLRECLPLIVFLRNRLKYALTGDE VKKICMQRFIKIDGKVRVDVTYPAGFMDVISIEKTGEHFRLVYDTKGRFAVHRITVEEAK YKLCKVRKITVGVKGIPHLVTHDARTIRYPDPVIKVNDTVQIDLGTGKIINFIKFDTGNL CMVIGGANLGRVGVITNRERHPGSFDVVHVKDANGNSFATRLSNIFVIGNGNKPWISLPR GKGIRLTVAEERDKRLATKQSSG |
| 767 |
P52888 (THOP1) |
 Estimated TM-score=0.918 |
>Thimet oligopeptidase (Length=689) MKPPAACAGDMADAASPCSVVNDLRWDLSAQQIEERTRELIEQTKRVYDQVGTQEFEDVS YESTLKALADVEVTYTVQRNILDFPQHVSPSKDIRTASTEADKKLSEFDVEMSMREDVYQ RIVWLQEKVQKDSLRPEAARYLERLIKLGRRNGLHLPRETQENIKRIKKKLSLLCIDFNK NLNEDTTFLPFTLQELGGLPEDFLNSLEKMEDGKLKVTLKYPHYFPLLKKCHVPETRRKV EEAFNCRCKEENCAILKELVTLRAQKSRLLGFHTHADYVLEMNMAKTSQTVATFLDELAQ KLKPLGEQERAVILELKRAECERRGLPFDGRIRAWDMRYYMNQVEETRYCVDQNLLKEYF PVQVVTHGLLGIYQELLGLAFHHEEGASAWHEDVRLYTARDAASGEVVGKFYLDLYPREG KYGHAACFGLQPGCLRQDGSRQIAIAAMVANFTKPTADAPSLLQHDEVETYFHEFGHVMH QLCSQAEFAMFSGTHVERDFVEAPSQMLENWVWEQEPLLRMSRHYRTGSAVPRELLEKLI ESRQANTGLFNLRQIVLAKVDQALHTQTDADPAEEYARLCQEILGVPATPGTNMPATFGH LAGGYDAQYYGYLWSEVYSMDMFHTRFKQEGVLNSKVGMDYRSCILRPGGSEDASAMLRR FLGRDPKQDAFLLSKGLQVGGCEPEPQVC |
| 768 |
Q9BRA2 (TXNDC17) |
 Estimated TM-score=0.918 |
>Thioredoxin domain-containing protein 17 (Length=123) MARYEEVSVSGFEEFHRAVEQHNGKTIFAYFTGSKDAGGKSWCPDCVQAEPVVREGLKHI SEGCVFIYCQVGEKPYWKDPNNDFRKNLKVTAVPTLLKYGTPQKLVESECLQANLVEMLF SED |
| 769 |
O75365 (PTP4A3) |
 Estimated TM-score=0.918 |
>Protein tyrosine phosphatase type IVA 3 (Length=173) MARMNRPAPVEVSYKHMRFLITHNPTNATLSTFIEDLKKYGATTVVRVCEVTYDKTPLEK DGITVVDWPFDDGAPPPGKVVEDWLSLVKAKFCEAPGSCVAVHCVAGLGRAPVLVALALI ESGMKYEDAIQFIRQKRRGAINSKQLTYLEKYRPKQRLRFKDPHTHKTRCCVM |
| 770 |
Q6XQN6 (NAPRT) |
 Estimated TM-score=0.918 |
>Nicotinate phosphoribosyltransferase (Length=538) MAAEQDPEARAAARPLLTDLYQATMALGYWRAGRARDAAEFELFFRRCPFGGAFALAAGL RDCVRFLRAFRLRDADVQFLASVLPPDTDPAFFEHLRALDCSEVTVRALPEGSLAFPGVP LLQVSGPLLVVQLLETPLLCLVSYASLVATNAARLRLIAGPEKRLLEMGLRRAQGPDGGL TASTYSYLGGFDSSSNVLAGQLRGVPVAGTLAHSFVTSFSGSEVPPDPMLAPAAGEGPGV DLAAKAQVWLEQVCAHLGLGVQEPHPGERAAFVAYALAFPRAFQGLLDTYSVWRSGLPNF LAVALALGELGYRAVGVRLDSGDLLQQAQEIRKVFRAAAAQFQVPWLESVLIVVSNNIDE EALARLAQEGSEVNVIGIGTSVVTCPQQPSLGGVYKLVAVGGQPRMKLTEDPEKQTLPGS KAAFRLLGSDGSPLMDMLQLAEEPVPQAGQELRVWPPGAQEPCTVRPAQVEPLLRLCLQQ GQLCEPLPSLAESRALAQLSLSRLSPEHRRLRSPAQYQVVLSERLQALVNSLCAGQSP |
| 771 |
P29033 (GJB2) |
 Estimated TM-score=0.918 |
>Gap junction beta-2 protein (Length=226) MDWGTLQTILGGVNKHSTSIGKIWLTVLFIFRIMILVVAAKEVWGDEQADFVCNTLQPGC KNVCYDHYFPISHIRLWALQLIFVSTPALLVAMHVAYRRHEKKRKFIKGEIKSEFKDIEE IKTQKVRIEGSLWWTYTSSIFFRVIFEAAFMYVFYVMYDGFSMQRLVKCNAWPCPNTVDC FVSRPTEKTVFTVFMIAVSGICILLNVTELCYLLIRYCSGKSKKPV |
| 772 |
Q9H0R8 (GABARAPL1) |
 Estimated TM-score=0.918 |
>Gamma-aminobutyric acid receptor-associated protein-like 1 (Length=117) MKFQYKEDHPFEYRKKEGEKIRKKYPDRVPVIVEKAPKARVPDLDKRKYLVPSDLTVGQF YFLIRKRIHLRPEDALFFFVNNTIPPTSATMGQLYEDNHEEDYFLYVAYSDESVYGK |
| 773 |
P41145 (OPRK1) |
 Estimated TM-score=0.918 |
>Kappa-type opioid receptor (Length=380) MDSPIQIFRGEPGPTCAPSACLPPNSSAWFPGWAEPDSNGSAGSEDAQLEPAHISPAIPV IITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYL MNSWPFGDVLCKIVISIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINI CIWLLSSSVGISAIVLGGTKVREDVDVIECSLQFPDDDYSWWDLFMKICVFIFAFVIPVL IIIVCYTLMILRLKSVRLLSGSREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALG STSHSTAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFPLKMRMERQSTSRV RNTVQDPAYLRDIDGMNKPV |
| 774 |
Q13889 (GTF2H3) |
 Estimated TM-score=0.917 |
>General transcription factor IIH subunit 3 (Length=308) MVSDEDELNLLVIVVDANPIWWGKQALKESQFTLSKCIDAVMVLGNSHLFMNRSNKLAVI ASHIQESRFLYPGKNGRLGDFFGDPGNPPEFNPSGSKDGKYELLTSANEVIVEEIKDLMT KSDIKGQHTETLLAGSLAKALCYIHRMNKEVKDNQEMKSRILVIKAAEDSALQYMNFMNV IFAAQKQNILIDACVLDSDSGLLQQACDITGGLYLKVPQMPSLLQYLLWVFLPDQDQRSQ LILPPPVHVDYRAACFCHRNLIEIGYVCSVCLSIFCNFSPICTTCETAFKISLPPVLKAK KKKLKVSA |
| 775 |
Q9UDW1 (UQCR10) |
 Estimated TM-score=0.917 |
>Cytochrome b-c1 complex subunit 9 (Length=63) MAAATLTSKLYSLLFRRTSTFALTIIVGVMFFERAFDQGADAIYDHINEGKLWKHIKHKY ENK |
| 776 |
Q5JS54 (PSMG4) |
 Estimated TM-score=0.917 |
>Proteasome assembly chaperone 4 (Length=123) MEGLVVAAGGDVSLHNFSARLWEQLVHFHVMRLTDSLFLWVGATPHLRNLAVAMCSRYDS IPVSTSLLGDTSDTTSTGLAQRLARKTNKQVFVSYNLQNTDSNFALLVENRIKEEMEAFP EKF |
| 777 |
P60709 (ACTB) |
 Estimated TM-score=0.917 |
>Actin, cytoplasmic 1 (Length=375) MDDDIAALVVDNGSGMCKAGFAGDDAPRAVFPSIVGRPRHQGVMVGMGQKDSYVGDEAQS KRGILTLKYPIEHGIVTNWDDMEKIWHHTFYNELRVAPEEHPVLLTEAPLNPKANREKMT QIMFETFNTPAMYVAIQAVLSLYASGRTTGIVMDSGDGVTHTVPIYEGYALPHAILRLDL AGRDLTDYLMKILTERGYSFTTTAEREIVRDIKEKLCYVALDFEQEMATAASSSSLEKSY ELPDGQVITIGNERFRCPEALFQPSFLGMESCGIHETTFNSIMKCDVDIRKDLYANTVLS GGTTMYPGIADRMQKEITALAPSTMKIKIIAPPERKYSVWIGGSILASLSTFQQMWISKQ EYDESGPSIVHRKCF |
| 778 |
P62249 (RPS16) |
 Estimated TM-score=0.917 |
>40S ribosomal protein S16 (Length=146) MPSKGPLQSVQVFGRKKTATAVAHCKRGNGLIKVNGRPLEMIEPRTLQYKLLEPVLLLGK ERFAGVDIRVRVKGGGHVAQIYAIRQSISKALVAYYQKYVDEASKKEIKDILIQYDRTLL VADPRRCESKKFGGPGARARYQKSYR |
| 779 |
P62899 (RPL31) |
 Estimated TM-score=0.917 |
>60S ribosomal protein L31 (Length=125) MAPAKKGGEKKKGRSAINEVVTREYTINIHKRIHGVGFKKRAPRALKEIRKFAMKEMGTP DVRIDTRLNKAVWAKGIRNVPYRIRVRLSRKRNEDEDSPNKLYTLVTYVPVTTFKNLQTV NVDEN |
| 780 |
Q96QV6 (H2AC1) |
 Estimated TM-score=0.916 |
>Histone H2A type 1-A (Length=131) MSGRGKQGGKARAKSKSRSSRAGLQFPVGRIHRLLRKGNYAERIGAGAPVYLAAVLEYLT AEILELAGNASRDNKKTRIIPRHLQLAIRNDEELNKLLGGVTIAQGGVLPNIQAVLLPKK TESHHHKAQSK |
| 781 |
P07355 (ANXA2) |
 Estimated TM-score=0.916 |
>Annexin A2 (Length=339) MSTVHEILCKLSLEGDHSTPPSAYGSVKAYTNFDAERDALNIETAIKTKGVDEVTIVNIL TNRSNAQRQDIAFAYQRRTKKELASALKSALSGHLETVILGLLKTPAQYDASELKASMKG LGTDEDSLIEIICSRTNQELQEINRVYKEMYKTDLEKDIISDTSGDFRKLMVALAKGRRA EDGSVIDYELIDQDARDLYDAGVKRKGTDVPKWISIMTERSVPHLQKVFDRYKSYSPYDM LESIRKEVKGDLENAFLNLVQCIQNKPLYFADRLYDSMKGKGTRDKVLIRIMVSRSEVDM LKIRSEFKRKYGKSLYYYIQQDTKGDYQKALLYLCGGDD |
| 782 |
P10632 (CYP2C8) |
 Estimated TM-score=0.916 |
>Cytochrome P450 2C8 (Length=490) MEPFVVLVLCLSFMLLFSLWRQSCRRRKLPPGPTPLPIIGNMLQIDVKDICKSFTNFSKV YGPVFTVYFGMNPIVVFHGYEAVKEALIDNGEEFSGRGNSPISQRITKGLGIISSNGKRW KEIRRFSLTTLRNFGMGKRSIEDRVQEEAHCLVEELRKTKASPCDPTFILGCAPCNVICS VVFQKRFDYKDQNFLTLMKRFNENFRILNSPWIQVCNNFPLLIDCFPGTHNKVLKNVALT RSYIREKVKEHQASLDVNNPRDFIDCFLIKMEQEKDNQKSEFNIENLVGTVADLFVAGTE TTSTTLRYGLLLLLKHPEVTAKVQEEIDHVIGRHRSPCMQDRSHMPYTDAVVHEIQRYSD LVPTGVPHAVTTDTKFRNYLIPKGTTIMALLTSVLHDDKEFPNPNIFDPGHFLDKNGNFK KSDYFMPFSAGKRICAGEGLARMELFLFLTTILQNFNLKSVDDLKNLNTTAVTKGIVSLP PSYQICFIPV |
| 783 |
O14732 (IMPA2) |
 Estimated TM-score=0.916 |
>Inositol monophosphatase 2 (Length=288) MKPSGEDQAALAAGPWEECFQAAVQLALRAGQIIRKALTEEKRVSTKTSAADLVTETDHL VEDLIISELRERFPSHRFIAEEAAASGAKCVLTHSPTWIIDPIDGTCNFVHRFPTVAVSI GFAVRQELEFGVIYHCTEERLYTGRRGRGAFCNGQRLRVSGETDLSKALVLTEIGPKRDP ATLKLFLSNMERLLHAKAHGVRVIGSSTLALCHLASGAADAYYQFGLHCWDLAAATVIIR EAGGIVIDTSGGPLDLMACRVVAASTREMAMLIAQALQTINYGRDDEK |
| 784 |
Q16718 (NDUFA5) |
 Estimated TM-score=0.916 |
>NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5 (Length=116) MAGVLKKTTGLVGLAVCNTPHERLRILYTKILDVLEEIPKNAAYRKYTEQITNEKLAMVK AEPDVKKLEDQLQGGQLEEVILQAEHELNLARKMREWKLWEPLVEEPPADQWKWPI |
| 785 |
O95257 (GADD45G) |
 Estimated TM-score=0.916 |
>Growth arrest and DNA damage-inducible protein GADD45 gamma (Length=159) MTLEEVRGQDTVPESTARMQGAGKALHELLLSAQRQGCLTAGVYESAKVLNVDPDNVTFC VLAAGEEDEGDIALQIHFTLIQAFCCENDIDIVRVGDVQRLAAIVGAGEEAGAPGDLHCI LISNPNEDAWKDPALEKLSLFCEESRSVNDWVPSITLPE |
| 786 |
Q9NZD4 (AHSP) |
 Estimated TM-score=0.916 |
>Alpha-hemoglobin-stabilizing protein (Length=102) MALLKANKDLISAGLKEFSVLLNQQVFNDPLVSEEDMVTVVEDWMNFYINYYRQQVTGEP QERDKALQELRQELNTLANPFLAKYRDFLKSHELPSHPPPSS |
| 787 |
Q5MNZ6 (WDR45B) |
 Estimated TM-score=0.915 |
>WD repeat domain phosphoinositide-interacting protein 3 (Length=344) MNLLPCNPHGNGLLYAGFNQDHGCFACGMENGFRVYNTDPLKEKEKQEFLEGGVGHVEML FRCNYLALVGGGKKPKYPPNKVMIWDDLKKKTVIEIEFSTEVKAVKLRRDRIVVVLDSMI KVFTFTHNPHQLHVFETCYNPKGLCVLCPNSNNSLLAFPGTHTGHVQLVDLASTEKPPVD IPAHEGVLSCIALNLQGTRIATASEKGTLIRIFDTSSGHLIQELRRGSQAANIYCINFNQ DASLICVSSDHGTVHIFAAEDPKRNKQSSLASASFLPKYFSSKWSFSKFQVPSGSPCICA FGTEPNAVIAICADGSYYKFLFNPKGECIRDVYAQFLEMTDDKL |
| 788 |
P57730 (CARD18) |
 Estimated TM-score=0.915 |
>Caspase recruitment domain-containing protein 18 (Length=90) MADQLLRKKRRIFIHSVGAGTINALLDCLLEDEVISQEDMNKVRDENDTVMDKARVLIDL VTGKGPKSCCKFIKHLCEEDPQLASKMGLH |
| 789 |
Q14019 (COTL1) |
 Estimated TM-score=0.915 |
>Coactosin-like protein (Length=142) MATKIDKEACRAAYNLVRDDGSAVIWVTFKYDGSTIVPGEQGAEYQHFIQQCTDDVRLFA FVRFTTGDAMSKRSKFALITWIGENVSGLQRAKTGTDKTLVKEVVQNFAKEFVISDRKEL EEDFIKSELKKAGGANYDAQTE |
| 790 |
Q8TBE9 (NANP) |
 Estimated TM-score=0.915 |
>N-acylneuraminate-9-phosphatase (Length=248) MGLSRVRAVFFDLDNTLIDTAGASRRGMLEVIKLLQSKYHYKEEAEIICDKVQVKLSKEC FHPYNTCITDLRTSHWEEAIQETKGGAANRKLAEECYFLWKSTRLQHMTLAEDVKAMLTE LRKEVRLLLLTNGDRQTQREKIEACACQSYFDAVVVGGEQREEKPAPSIFYYCCNLLGVQ PGDCVMVGDTLETDIQGGLNAGLKATVWINKNGIVPLKSSPVPHYMVSSVLELPALLQSI DCKVSMST |
| 791 |
Q9UDX3 (SEC14L4) |
 Estimated TM-score=0.915 |
>SEC14-like protein 4 (Length=406) MSSRVGDLSPQQQEALARFRENLQDLLPILPNADDYFLLRWLRARNFDLQKSEDMLRRHM EFRKQQDLDNIVTWQPPEVIQLYDSGGLCGYDYEGCPVYFNIIGSLDPKGLLLSASKQDM IRKRIKVCELLLHECELQTQKLGRKIEMALMVFDMEGLSLKHLWKPAVEVYQQFFSILEA NYPETLKNLIVIRAPKLFPVAFNLVKSFMSEETRRKIVILGDNWKQELTKFISPDQLPVE FGGTMTDPDGNPKCLTKINYGGEVPKSYYLCEQVRLQYEHTRSVGRGSSLQVENEILFPG CVLRWQFASDGGDIGFGVFLKTKMGEQQSAREMTEVLPSQRYNAHMVPEDGSLTCLQAGV YVLRFDNTYSRMHAKKLSYTVEVLLPDKASEETLQSLKAMRPSPTQ |
| 792 |
O95298 (NDUFC2) |
 Estimated TM-score=0.915 |
>NADH dehydrogenase [ubiquinone] 1 subunit C2 (Length=119) MIARRNPEPLRFLPDEARSLPPPKLTDPRLLYIGFLGYCSGLIDNLIRRRPIATAGLHRQ LLYITAFFFAGYYLVKREDYLYAVRDREMFGYMKLHPEDFPEEDKKTYGEIFEKFHPIR |
| 793 |
Q13084 (MRPL28) |
 Estimated TM-score=0.915 |
>39S ribosomal protein L28, mitochondrial (Length=256) MPLHKYPVWLWKRLQLREGICSRLPGHYLRSLEEERTPTPVHYRPHGAKFKINPKNGQRE RVEDVPIPIYFPPESQRGLWGGEGWILGQIYANNDKLSKRLKKVWKPQLFEREFYSEILD KKFTVTVTMRTLDLIDEAYGLDFYILKTPKEDLCSKFGMDLKRGMLLRLARQDPQLHPED PERRAAIYDKYKEFAIPEEEAEWVGLTLEEAIEKQRLLEEKDPVPLFKIYVAELIQQLQQ QALSEPAVVQKRASGQ |
| 794 |
P01040 (CSTA) |
 Estimated TM-score=0.915 |
>Cystatin-A (Length=98) MIPGGLSEAKPATPEIQEIVDKVKPQLEEKTNETYGKLEAVQYKTQVVAGTNYYIKVRAG DNKYMHLKVFKSLPGQNEDLVLTGYQVDKNKDDELTGF |
| 795 |
Q9BZE1 (MRPL37) |
 Estimated TM-score=0.915 |
>39S ribosomal protein L37, mitochondrial (Length=423) MALASGPARRALAGSGQLGLGGFGAPRRGAYEWGVRSTRKSEPPPLDRVYEIPGLEPITF AGKMHFVPWLARPIFPPWDRGYKDPRFYRSPPLHEHPLYKDQACYIFHHRCRLLEGVKQA LWLTKTKLIEGLPEKVLSLVDDPRNHIENQDECVLNVISHARLWQTTEEIPKRETYCPVI VDNLIQLCKSQILKHPSLARRICVQNSTFSATWNRESLLLQVRGSGGARLSTKDPLPTIA SREEIEATKNHVLETFYPISPIIDLHECNIYDVKNDTGFQEGYPYPYPHTLYLLDKANLR PHRLQPDQLRAKMILFAFGSALAQARLLYGNDAKVLEQPVVVQSVGTDGRVFHFLVFQLN TTDLDCNEGVKNLAWVDSDQLLYQHFWCLPVIKKRVVVEPVGPVGFKPETFRKFLALYLH GAA |
| 796 |
P35813 (PPM1A) |
 Estimated TM-score=0.915 |
>Protein phosphatase 1A (Length=382) MGAFLDKPKMEKHNAQGQGNGLRYGLSSMQGWRVEMEDAHTAVIGLPSGLESWSFFAVYD GHAGSQVAKYCCEHLLDHITNNQDFKGSAGAPSVENVKNGIRTGFLEIDEHMRVMSEKKH GADRSGSTAVGVLISPQHTYFINCGDSRGLLCRNRKVHFFTQDHKPSNPLEKERIQNAGG SVMIQRVNGSLAVSRALGDFDYKCVHGKGPTEQLVSPEPEVHDIERSEEDDQFIILACDG IWDVMGNEELCDFVRSRLEVTDDLEKVCNEVVDTCLYKGSRDNMSVILICFPNAPKVSPE AVKKEAELDKYLECRVEEIIKKQGEGVPDLVHVMRTLASENIPSLPPGGELASKRNVIEA VYNRLNPYKNDDTDSTSTDDMW |
| 797 |
P10619 (CTSA) |
 Estimated TM-score=0.915 |
>Lysosomal protective protein (Length=480) MIRAAPPPLFLLLLLLLLLVSWASRGEAAPDQDEIQRLPGLAKQPSFRQYSGYLKGSGSK HLHYWFVESQKDPENSPVVLWLNGGPGCSSLDGLLTEHGPFLVQPDGVTLEYNPYSWNLI ANVLYLESPAGVGFSYSDDKFYATNDTEVAQSNFEALQDFFRLFPEYKNNKLFLTGESYA GIYIPTLAVLVMQDPSMNLQGLAVGNGLSSYEQNDNSLVYFAYYHGLLGNRLWSSLQTHC CSQNKCNFYDNKDLECVTNLQEVARIVGNSGLNIYNLYAPCAGGVPSHFRYEKDTVVVQD LGNIFTRLPLKRMWHQALLRSGDKVRMDPPCTNTTAASTYLNNPYVRKALNIPEQLPQWD MCNFLVNLQYRRLYRSMNSQYLKLLSSQKYQILLYNGDVDMACNFMGDEWFVDSLNQKME VQRRPWLVKYGDSGEQIAGFVKEFSHIAFLTIKGAGHMVPTDKPLAAFTMFSRFLNKQPY |
| 798 |
P23368 (ME2) |
 Estimated TM-score=0.914 |
>NAD-dependent malic enzyme, mitochondrial (Length=584) MLSRLRVVSTTCTLACRHLHIKEKGKPLMLNPRTNKGMAFTLQERQMLGLQGLLPPKIET QDIQALRFHRNLKKMTSPLEKYIYIMGIQERNEKLFYRILQDDIESLMPIVYTPTVGLAC SQYGHIFRRPKGLFISISDRGHVRSIVDNWPENHVKAVVVTDGERILGLGDLGVYGMGIP VGKLCLYTACAGIRPDRCLPVCIDVGTDNIALLKDPFYMGLYQKRDRTQQYDDLIDEFMK AITDRYGRNTLIQFEDFGNHNAFRFLRKYREKYCTFNDDIQGTAAVALAGLLAAQKVISK PISEHKILFLGAGEAALGIANLIVMSMVENGLSEQEAQKKIWMFDKYGLLVKGRKAKIDS YQEPFTHSAPESIPDTFEDAVNILKPSTIIGVAGAGRLFTPDVIRAMASINERPVIFALS NPTAQAECTAEEAYTLTEGRCLFASGSPFGPVKLTDGRVFTPGQGNNVYIFPGVALAVIL CNTRHISDSVFLEAAKALTSQLTDEELAQGRLYPPLANIQEVSINIAIKVTEYLYANKMA FRYPEPEDKAKYVKERTWRSEYDSLLPDVYEWPESASSPPVITE |
| 799 |
Q15109 (AGER) |
 Estimated TM-score=0.914 |
>Advanced glycosylation end product-specific receptor (Length=404) MAAGTAVGAWVLVLSLWGAVVGAQNITARIGEPLVLKCKGAPKKPPQRLEWKLNTGRTEA WKVLSPQGGGPWDSVARVLPNGSLFLPAVGIQDEGIFRCQAMNRNGKETKSNYRVRVYQI PGKPEIVDSASELTAGVPNKVGTCVSEGSYPAGTLSWHLDGKPLVPNEKGVSVKEQTRRH PETGLFTLQSELMVTPARGGDPRPTFSCSFSPGLPRHRALRTAPIQPRVWEPVPLEEVQL VVEPEGGAVAPGGTVTLTCEVPAQPSPQIHWMKDGVPLPLPPSPVLILPEIGPQDQGTYS CVATHSSHGPQESRAVSISIIEPGEEGPTAGSVGGSGLGTLALALGILGGLGTAALLIGV ILWQRRQRRGEERKAPENQEEEEERAELNQSEEPEAGESSTGGP |
| 800 |
Q9GZT4 (SRR) |
 Estimated TM-score=0.914 |
>Serine racemase (Length=340) MCAQYCISFADVEKAHINIRDSIHLTPVLTSSILNQLTGRNLFFKCELFQKTGSFKIRGA LNAVRSLVPDALERKPKAVVTHSSGNHGQALTYAAKLEGIPAYIVVPQTAPDCKKLAIQA YGASIVYCEPSDESRENVAKRVTEETEGIMVHPNQEPAVIAGQGTIALEVLNQVPLVDAL VVPVGGGGMLAGIAITVKALKPSVKVYAAEPSNADDCYQSKLKGKLMPNLYPPETIADGV KSSIGLNTWPIIRDLVDDIFTVTEDEIKCATQLVWERMKLLIEPTAGVGVAAVLSQHFQT VSPEVKNICIVLSGGNVDLTSSITWVKQAERPASYQSVSV |
[an error occurred while processing this directive]
yangzhanglab![]() umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218