


Go to page (17 pages in total): [First page] << < 1 2 3 4 5 6 7 8 9 10 > >> [Last page] [All entries in one page].
| # | Uniprot ID | D-I-TASSER-AF2 models ranked by estimated TM-score (click to view) |
Full name and sequence |
| 401 |
P00492 (HPRT1) |
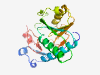 Estimated TM-score=0.947 |
>Hypoxanthine-guanine phosphoribosyltransferase (Length=218) MATRSPGVVISDDEPGYDLDLFCIPNHYAEDLERVFIPHGLIMDRTERLARDVMKEMGGH HIVALCVLKGGYKFFADLLDYIKALNRNSDRSIPMTVDFIRLKSYCNDQSTGDIKVIGGD DLSTLTGKNVLIVEDIIDTGKTMQTLLSLVRQYNPKMVKVASLLVKRTPRSVGYKPDFVG FEIPDKFVVGYALDYNEYFRDLNHVCVISETGKAKYKA |
| 402 |
O15540 (FABP7) |
 Estimated TM-score=0.947 |
>Fatty acid-binding protein, brain (Length=132) MVEAFCATWKLTNSQNFDEYMKALGVGFATRQVGNVTKPTVIISQEGDKVVIRTLSTFKN TEISFQLGEEFDETTADDRNCKSVVSLDGDKLVHIQKWDGKETNFVREIKDGKMVMTLTF GDVVAVRHYEKA |
| 403 |
Q8N9L9 (ACOT4) |
 Estimated TM-score=0.947 |
>Peroxisomal succinyl-coenzyme A thioesterase (Length=421) MSATLILEPPGRCCWNEPVRIAVRGLAPEQRVTLRASLRDEKGALFRAHARYCADARGEL DLERAPALGGSFAGLEPMGLLWALEPEKPFWRFLKRDVQIPFVVELEVLDGHDPEPGRLL CQAQHERHFLPPGVRRQSVRAGRVRATLFLPPGPGPFPGIIDIFGIGGGLLEYRASLLAG HGFATLALAYYNFEDLPNNMDNISLEYFEEAVCYMLQHPQVKGPGIGLLGISLGADICLS MASFLKNVSATVSINGSGISGNTAINYKHSSIPPLGYDLRRIKVAFSGLVDIVDIRNALV GGYKNPSMIPIEKAQGPILLIVGQDDHNWRSELYAQTVSERLQAHGKEKPQIICYPGTGH YIEPPYFPLCPASLHRLLNKHVIWGGEPRAHSKAQEDAWKQILAFFCKHLGGTQKTAVPK L |
| 404 |
Q96DE0 (NUDT16) |
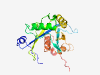 Estimated TM-score=0.946 |
>U8 snoRNA-decapping enzyme (Length=195) MAGARRLELGEALALGSGWRHACHALLYAPDPGMLFGRIPLRYAILMQMRFDGRLGFPGG FVDTQDRSLEDGLNRELREELGEAAAAFRVERTDYRSSHVGSGPRVVAHFYAKRLTLEEL LAVEAGATRAKDHGLEVLGLVRVPLYTLRDGVGGLPTFLENSFIGSAREQLLEALQDLGL LQSGSISGLKIPAHH |
| 405 |
P07737 (PFN1) |
 Estimated TM-score=0.946 |
>Profilin-1 (Length=140) MAGWNAYIDNLMADGTCQDAAIVGYKDSPSVWAAVPGKTFVNITPAEVGVLVGKDRSSFY VNGLTLGGQKCSVIRDSLLQDGEFSMDLRTKSTGGAPTFNVTVTKTDKTLVLLMGKEGVH GGLINKKCYEMASHLRRSQY |
| 406 |
O60234 (GMFG) |
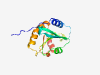 Estimated TM-score=0.946 |
>Glia maturation factor gamma (Length=142) MSDSLVVCEVDPELTEKLRKFRFRKETDNAAIIMKVDKDRQMVVLEEEFQNISPEELKME LPERQPRFVVYSYKYVHDDGRVSYPLCFIFSSPVGCKPEQQMMYAGSKNRLVQTAELTKV FEIRTTDDLTEAWLQEKLSFFR |
| 407 |
Q7Z4W1 (DCXR) |
 Estimated TM-score=0.946 |
>L-xylulose reductase (Length=244) MELFLAGRRVLVTGAGKGIGRGTVQALHATGARVVAVSRTQADLDSLVRECPGIEPVCVD LGDWEATERALGSVGPVDLLVNNAAVALLQPFLEVTKEAFDRSFEVNLRAVIQVSQIVAR GLIARGVPGAIVNVSSQCSQRAVTNHSVYCSTKGALDMLTKVMALELGPHKIRVNAVNPT VVMTSMGQATWSDPHKAKTMLNRIPLGKFAEVEHVVNAILFLLSDRSGMTTGSTLPVEGG FWAC |
| 408 |
Q9Y2Q5 (LAMTOR2) |
 Estimated TM-score=0.946 |
>Ragulator complex protein LAMTOR2 (Length=125) MLRPKALTQVLSQANTGGVQSTLLLNNEGSLLAYSGYGDTDARVTAAIASNIWAAYDRNG NQAFNEDNLKFILMDCMEGRVAITRVANLLLCMYAKETVGFGMLKAKAQALVQYLEEPLT QVAAS |
| 409 |
P02753 (RBP4) |
 Estimated TM-score=0.946 |
>Retinol-binding protein 4 (Length=201) MKWVWALLLLAALGSGRAERDCRVSSFRVKENFDKARFSGTWYAMAKKDPEGLFLQDNIV AEFSVDETGQMSATAKGRVRLLNNWDVCADMVGTFTDTEDPAKFKMKYWGVASFLQKGND DHWIVDTDYDTYAVQYSCRLLNLDGTCADSYSFVFSRDPNGLPPEAQKIVRQRQEELCLA RQYRLIVHNGYCDGRSERNLL |
| 410 |
P22061 (PCMT1) |
 Estimated TM-score=0.946 |
>Protein-L-isoaspartate(D-aspartate) O-methyltransferase (Length=227) MAWKSGGASHSELIHNLRKNGIIKTDKVFEVMLATDRSHYAKCNPYMDSPQSIGFQATIS APHMHAYALELLFDQLHEGAKALDVGSGSGILTACFARMVGCTGKVIGIDHIKELVDDSV NNVRKDDPTLLSSGRVQLVVGDGRMGYAEEAPYDAIHVGAAAPVVPQALIDQLKPGGRLI LPVGPAGGNQMLEQYDKLQDGSIKMKPLMGVIYVPLTDKEKQWSRWK |
| 411 |
P07360 (C8G) |
 Estimated TM-score=0.946 |
>Complement component C8 gamma chain (Length=202) MLPPGTATLLTLLLAAGSLGQKPQRPRRPASPISTIQPKANFDAQQFAGTWLLVAVGSAC RFLQEQGHRAEATTLHVAPQGTAMAVSTFRKLDGICWQVRQLYGDTGVLGRFLLQARDAR GAVHVVVAETDYQSFAVLYLERAGQLSVKLYARSLPVSDSVLSGFEQRVQEAHLTEDQIF YFPKYGFCEAADQFHVLDEVRR |
| 412 |
Q9UHA7 (IL36A) |
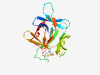 Estimated TM-score=0.946 |
>Interleukin-36 alpha (Length=158) MEKALKIDTPQQGSIQDINHRVWVLQDQTLIAVPRKDRMSPVTIALISCRHVETLEKDRG NPIYLGLNGLNLCLMCAKVGDQPTLQLKEKDIMDLYNQPEPVKSFLFYHSQSGRNSTFES VAFPGWFIAVSSEGGCPLILTQELGKANTTDFGLTMLF |
| 413 |
P40429 (RPL13A) |
 Estimated TM-score=0.946 |
>60S ribosomal protein L13a (Length=203) MAEVQVLVLDGRGHLLGRLAAIVAKQVLLGRKVVVVRCEGINISGNFYRNKLKYLAFLRK RMNTNPSRGPYHFRAPSRIFWRTVRGMLPHKTKRGQAALDRLKVFDGIPPPYDKKKRMVV PAALKVVRLKPTRKFAYLGRLAHEVGWKYQAVTATLEEKRKEKAKIHYRKKKQLMRLRKQ AEKNVEKKIDKYTEVLKTHGLLV |
| 414 |
O43617 (TRAPPC3) |
 Estimated TM-score=0.946 |
>Trafficking protein particle complex subunit 3 (Length=180) MSRQANRGTESKKMSSELFTLTYGALVTQLCKDYENDEDVNKQLDKMGFNIGVRLIEDFL ARSNVGRCHDFRETADVIAKVAFKMYLGITPSITNWSPAGDEFSLILENNPLVDFVELPD NHSSLIYSNLLCGVLRGALEMVQMAVEAKFVQDTLKGDGVTEIRMRFIRRIEDNLPAGEE |
| 415 |
Q9NP95 (FGF20) |
 Estimated TM-score=0.946 |
>Fibroblast growth factor 20 (Length=211) MAPLAEVGGFLGGLEGLGQQVGSHFLLPPAGERPPLLGERRSAAERSARGGPGAAQLAHL HGILRRRQLYCRTGFHLQILPDGSVQGTRQDHSLFGILEFISVAVGLVSIRGVDSGLYLG MNDKGELYGSEKLTSECIFREQFEENWYNTYSSNIYKHGDTGRRYFVALNKDGTPRDGAR SKRHQKFTHFLPRPVDPERVPELYKDLLMYT |
| 416 |
Q9NV96 (TMEM30A) |
 Estimated TM-score=0.946 |
>Cell cycle control protein 50A (Length=361) MAMNYNAKDEVDGGPPCAPGGTAKTRRPDNTAFKQQRLPAWQPILTAGTVLPIFFIIGLI FIPIGIGIFVTSNNIREIEIDYTGTEPSSPCNKCLSPDVTPCFCTINFTLEKSFEGNVFM YYGLSNFYQNHRRYVKSRDDSQLNGDSSALLNPSKECEPYRRNEDKPIAPCGAIANSMFN DTLELFLIGNDSYPIPIALKKKGIAWWTDKNVKFRNPPGGDNLEERFKGTTKPVNWLKPV YMLDSDPDNNGFINEDFIVWMRTAALPTFRKLYRLIERKSDLHPTLPAGRYSLNVTYNYP VHYFDGRKRMILSTISWMGGKNPFLGIAYIAVGSISFLLGVVLLVINHKYRNSSNTADIT I |
| 417 |
P22304 (IDS) |
 Estimated TM-score=0.946 |
>Iduronate 2-sulfatase (Length=550) MPPPRTGRGLLWLGLVLSSVCVALGSETQANSTTDALNVLLIIVDDLRPSLGCYGDKLVR SPNIDQLASHSLLFQNAFAQQAVCAPSRVSFLTGRRPDTTRLYDFNSYWRVHAGNFSTIP QYFKENGYVTMSVGKVFHPGISSNHTDDSPYSWSFPPYHPSSEKYENTKTCRGPDGELHA NLLCPVDVLDVPEGTLPDKQSTEQAIQLLEKMKTSASPFFLAVGYHKPHIPFRYPKEFQK LYPLENITLAPDPEVPDGLPPVAYNPWMDIRQREDVQALNISVPYGPIPVDFQRKIRQSY FASVSYLDTQVGRLLSALDDLQLANSTIIAFTSDHGWALGEHGEWAKYSNFDVATHVPLI FYVPGRTASLPEAGEKLFPYLDPFDSASQLMEPGRQSMDLVELVSLFPTLAGLAGLQVPP RCPVPSFHVELCREGKNLLKHFRFRDLEEDPYLPGNPRELIAYSQYPRPSDIPQWNSDKP SLKDIKIMGYSIRTIDYRYTVWVGFNPDEFLANFSDIHAGELYFVDSDPLQDHNMYNDSQ GGDLFQLLMP |
| 418 |
Q9BR01 (SULT4A1) |
 Estimated TM-score=0.946 |
>Sulfotransferase 4A1 (Length=284) MAESEAETPSTPGEFESKYFEFHGVRLPPFCRGKMEEIANFPVRPSDVWIVTYPKSGTSL LQEVVYLVSQGADPDEIGLMNIDEQLPVLEYPQPGLDIIKELTSPRLIKSHLPYRFLPSD LHNGDSKVIYMARNPKDLVVSYYQFHRSLRTMSYRGTFQEFCRRFMNDKLGYGSWFEHVQ EFWEHRMDSNVLFLKYEDMHRDLVTMVEQLARFLGVSCDKAQLEALTEHCHQLVDQCCNA EALPVGRGRVGLWKDIFTVSMNEKFDLVYKQKMGKCDLTFDFYL |
| 419 |
Q01469 (FABP5) |
 Estimated TM-score=0.946 |
>Fatty acid-binding protein 5 (Length=135) MATVQQLEGRWRLVDSKGFDEYMKELGVGIALRKMGAMAKPDCIITCDGKNLTIKTESTL KTTQFSCTLGEKFEETTADGRKTQTVCNFTDGALVQHQEWDGKESTITRKLKDGKLVVEC VMNNVTCTRIYEKVE |
| 420 |
P25788 (PSMA3) |
 Estimated TM-score=0.946 |
>Proteasome subunit alpha type-3 (Length=255) MSSIGTGYDLSASTFSPDGRVFQVEYAMKAVENSSTAIGIRCKDGVVFGVEKLVLSKLYE EGSNKRLFNVDRHVGMAVAGLLADARSLADIAREEASNFRSNFGYNIPLKHLADRVAMYV HAYTLYSAVRPFGCSFMLGSYSVNDGAQLYMIDPSGVSYGYWGCAIGKARQAAKTEIEKL QMKEMTCRDIVKEVAKIIYIVHDEVKDKAFELELSWVGELTNGRHEIVPKDIREEAEKYA KESLKEEDESDDDNM |
| 421 |
P22033 (MMUT) |
 Estimated TM-score=0.946 |
>Methylmalonyl-CoA mutase, mitochondrial (Length=750) MLRAKNQLFLLSPHYLRQVKESSGSRLIQQRLLHQQQPLHPEWAALAKKQLKGKNPEDLI WHTPEGISIKPLYSKRDTMDLPEELPGVKPFTRGPYPTMYTFRPWTIRQYAGFSTVEESN KFYKDNIKAGQQGLSVAFDLATHRGYDSDNPRVRGDVGMAGVAIDTVEDTKILFDGIPLE KMSVSMTMNGAVIPVLANFIVTGEEQGVPKEKLTGTIQNDILKEFMVRNTYIFPPEPSMK IIADIFEYTAKHMPKFNSISISGYHMQEAGADAILELAYTLADGLEYSRTGLQAGLTIDE FAPRLSFFWGIGMNFYMEIAKMRAGRRLWAHLIEKMFQPKNSKSLLLRAHCQTSGWSLTE QDPYNNIVRTAIEAMAAVFGGTQSLHTNSFDEALGLPTVKSARIARNTQIIIQEESGIPK VADPWGGSYMMECLTNDVYDAALKLINEIEEMGGMAKAVAEGIPKLRIEECAARRQARID SGSEVIVGVNKYQLEKEDAVEVLAIDNTSVRNRQIEKLKKIKSSRDQALAERCLAALTEC AASGDGNILALAVDASRARCTVGEITDALKKVFGEHKANDRMVSGAYRQEFGESKEITSA IKRVHKFMEREGRRPRLLVAKMGQDGHDRGAKVIATGFADLGFDVDIGPLFQTPREVAQQ AVDADVHAVGISTLAAGHKTLVPELIKELNSLGRPDILVMCGGVIPPQDYEFLFEVGVSN VFGPGTRIPKAAVQVLDDIEKCLEKKQQSV |
| 422 |
P27338 (MAOB) |
 Estimated TM-score=0.946 |
>Amine oxidase [flavin-containing] B (Length=520) MSNKCDVVVVGGGISGMAAAKLLHDSGLNVVVLEARDRVGGRTYTLRNQKVKYVDLGGSY VGPTQNRILRLAKELGLETYKVNEVERLIHHVKGKSYPFRGPFPPVWNPITYLDHNNFWR TMDDMGREIPSDAPWKAPLAEEWDNMTMKELLDKLCWTESAKQLATLFVNLCVTAETHEV SALWFLWYVKQCGGTTRIISTTNGGQERKFVGGSGQVSERIMDLLGDRVKLERPVIYIDQ TRENVLVETLNHEMYEAKYVISAIPPTLGMKIHFNPPLPMMRNQMITRVPLGSVIKCIVY YKEPFWRKKDYCGTMIIDGEEAPVAYTLDDTKPEGNYAAIMGFILAHKARKLARLTKEER LKKLCELYAKVLGSLEALEPVHYEEKNWCEEQYSGGCYTTYFPPGILTQYGRVLRQPVDR IYFAGTETATHWSGYMEGAVEAGERAAREILHAMGKIPEDEIWQSEPESVDVPAQPITTT FLERHLPSVPGLLRLIGLTTIFSATALGFLAHKRGLLVRV |
| 423 |
Q9Y2S2 (CRYL1) |
 Estimated TM-score=0.946 |
>Lambda-crystallin homolog (Length=319) MASSAAGCVVIVGSGVIGRSWAMLFASGGFQVKLYDIEQQQIRNALENIRKEMKLLEQAG SLKGSLSVEEQLSLISGCPNIQEAVEGAMHIQECVPEDLELKKKIFAQLDSIIDDRVILS SSTSCLMPSKLFAGLVHVKQCIVAHPVNPPYYIPLVELVPHPETAPTTVDRTHALMKKIG QCPMRVQKEVAGFVLNRLQYAIISEAWRLVEEGIVSPSDLDLVMSEGLGMRYAFIGPLET MHLNAEGMLSYCDRYSEGIKHVLQTFGPIPEFSRATAEKVNQDMCMKVPDDPEHLAARRQ WRDECLMRLAKLKSQVQPQ |
| 424 |
Q6UWP2 (DHRS11) |
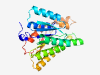 Estimated TM-score=0.946 |
>Dehydrogenase/reductase SDR family member 11 (Length=260) MARPGMERWRDRLALVTGASGGIGAAVARALVQQGLKVVGCARTVGNIEELAAECKSAGY PGTLIPYRCDLSNEEDILSMFSAIRSQHSGVDICINNAGLARPDTLLSGSTSGWKDMFNV NVLALSICTREAYQSMKERNVDDGHIININSMSGHRVLPLSVTHFYSATKYAVTALTEGL RQELREAQTHIRATCISPGVVETQFAFKLHDKDPEKAAATYEQMKCLKPEDVAEAVIYVL STPAHIQIGDIQMRPTEQVT |
| 425 |
P09382 (LGALS1) |
 Estimated TM-score=0.945 |
>Galectin-1 (Length=135) MACGLVASNLNLKPGECLRVRGEVAPDAKSFVLNLGKDSNNLCLHFNPRFNAHGDANTIV CNSKDGGAWGTEQREAVFPFQPGSVAEVCITFDQANLTVKLPDGYEFKFPNRLNLEAINY MAADGDFKIKCVAFD |
| 426 |
P51149 (RAB7A) |
 Estimated TM-score=0.945 |
>Ras-related protein Rab-7a (Length=207) MTSRKKVLLKVIILGDSGVGKTSLMNQYVNKKFSNQYKATIGADFLTKEVMVDDRLVTMQ IWDTAGQERFQSLGVAFYRGADCCVLVFDVTAPNTFKTLDSWRDEFLIQASPRDPENFPF VVLGNKIDLENRQVATKRAQAWCYSKNNIPYFETSAKEAINVEQAFQTIARNALKQETEV ELYNEFPEPIKLDKNDRAKASAESCSC |
| 427 |
P15153 (RAC2) |
 Estimated TM-score=0.945 |
>Ras-related C3 botulinum toxin substrate 2 (Length=192) MQAIKCVVVGDGAVGKTCLLISYTTNAFPGEYIPTVFDNYSANVMVDSKPVNLGLWDTAG QEDYDRLRPLSYPQTDVFLICFSLVSPASYENVRAKWFPEVRHHCPSTPIILVGTKLDLR DDKDTIEKLKEKKLAPITYPQGLALAKEIDSVKYLECSALTQRGLKTVFDEAIRAVLCPQ PTRQQKRACSLL |
| 428 |
P14384 (CPM) |
 Estimated TM-score=0.945 |
>Carboxypeptidase M (Length=443) MDFPCLWLGLLLPLVAALDFNYHRQEGMEAFLKTVAQNYSSVTHLHSIGKSVKGRNLWVL VVGRFPKEHRIGIPEFKYVANMHGDETVGRELLLHLIDYLVTSDGKDPEITNLINSTRIH IMPSMNPDGFEAVKKPDCYYSIGRENYNQYDLNRNFPDAFEYNNVSRQPETVAVMKWLKT ETFVLSANLHGGALVASYPFDNGVQATGALYSRSLTPDDDVFQYLAHTYASRNPNMKKGD ECKNKMNFPNGVTNGYSWYPLQGGMQDYNYIWAQCFEITLELSCCKYPREEKLPSFWNNN KASLIEYIKQVHLGVKGQVFDQNGNPLPNVIVEVQDRKHICPYRTNKYGEYYLLLLPGSY IINVTVPGHDPHITKVIIPEKSQNFSALKKDILLPFQGQLDSIPVSNPSCPMIPLYRNLP DHSAATKPSLFLFLVSLLHIFFK |
| 429 |
P40261 (NNMT) |
 Estimated TM-score=0.945 |
>Nicotinamide N-methyltransferase (Length=264) MESGFTSKDTYLSHFNPRDYLEKYYKFGSRHSAESQILKHLLKNLFKIFCLDGVKGDLLI DIGSGPTIYQLLSACESFKEIVVTDYSDQNLQELEKWLKKEPEAFDWSPVVTYVCDLEGN RVKGPEKEEKLRQAVKQVLKCDVTQSQPLGAVPLPPADCVLSTLCLDAACPDLPTYCRAL RNLGSLLKPGGFLVIMDALKSSYYMIGEQKFSSLPLGREAVEAAVKEAGYTIEWFEVISQ SYSSTMANNEGLFSLVARKLSRPL |
| 430 |
Q9NSD9 (FARSB) |
 Estimated TM-score=0.945 |
>Phenylalanine--tRNA ligase beta subunit (Length=589) MPTVSVKRDLLFQALGRTYTDEEFDELCFEFGLELDEITSEKEIISKEQGNVKAAGASDV VLYKIDVPANRYDLLCLEGLVRGLQVFKERIKAPVYKRVMPDGKIQKLIITEETAKIRPF AVAAVLRNIKFTKDRYDSFIELQEKLHQNICRKRALVAIGTHDLDTLSGPFTYTAKRPSD IKFKPLNKTKEYTACELMNIYKTDNHLKHYLHIIENKPLYPVIYDSNGVVLSMPPIINGD HSRITVNTRNIFIECTGTDFTKAKIVLDIIVTMFSEYCENQFTVEAAEVVFPNGKSHTFP ELAYRKEMVRADLINKKVGIRETPENLAKLLTRMYLKSEVIGDGNQIEIEIPPTRADIIH ACDIVEDAAIAYGYNNIQMTLPKTYTIANQFPLNKLTELLRHDMAAAGFTEALTFALCSQ EDIADKLGVDISATKAVHISNPKTAEFQVARTTLLPGLLKTIAANRKMPLPLKLFEISDI VIKDSNTDVGAKNYRHLCAVYYNKNPGFEIIHGLLDRIMQLLDVPPGEDKGGYVIKASEG PAFFPGRCAEIFARGQSVGKLGVLHPDVITKFELTMPCSSLEINVGPFL |
| 431 |
P25787 (PSMA2) |
 Estimated TM-score=0.945 |
>Proteasome subunit alpha type-2 (Length=234) MAERGYSFSLTTFSPSGKLVQIEYALAAVAGGAPSVGIKAANGVVLATEKKQKSILYDER SVHKVEPITKHIGLVYSGMGPDYRVLVHRARKLAQQYYLVYQEPIPTAQLVQRVASVMQE YTQSGGVRPFGVSLLICGWNEGRPYLFQSDPSGAYFAWKATAMGKNYVNGKTFLEKRYNE DLELEDAIHTAILTLKESFEGQMTEDNIEVGICNEAGFRRLTPTEVKDYLAAIA |
| 432 |
Q13232 (NME3) |
 Estimated TM-score=0.945 |
>Nucleoside diphosphate kinase 3 (Length=169) MICLVLTIFANLFPAACTGAHERTFLAVKPDGVQRRLVGEIVRRFERKGFKLVALKLVQA SEELLREHYAELRERPFYGRLVKYMASGPVVAMVWQGLDVVRTSRALIGATNPADAPPGT IRGDFCIEVGKNLIHGSDSVESARREIALWFRADELLCWEDSAGHWLYE |
| 433 |
Q9ULA0 (DNPEP) |
 Estimated TM-score=0.945 |
>Aspartyl aminopeptidase (Length=475) MQVAMNGKARKEAVQTAAKELLKFVNRSPSPFHAVAECRNRLLQAGFSELKETEKWNIKP ESKYFMTRNSSTIIAFAVGGQYVPGNGFSLIGAHTDSPCLRVKRRSRRSQVGFQQVGVET YGGGIWSTWFDRDLTLAGRVIVKCPTSGRLEQQLVHVERPILRIPHLAIHLQRNINENFG PNTEMHLVPILATAIQEELEKGTPEPGPLNAVDERHHSVLMSLLCAHLGLSPKDIVEMEL CLADTQPAVLGGAYDEFIFAPRLDNLHSCFCALQALIDSCAGPGSLATEPHVRMVTLYDN EEVGSESAQGAQSLLTELVLRRISASCQHPTAFEEAIPKSFMISADMAHAVHPNYLDKHE ENHRPLFHKGPVIKVNSKQRYASNAVSEALIREVANKVKVPLQDLMVRNDTPCGTTIGPI LASRLGLRVLDLGSPQLAMHSIREMACTTGVLQTLTLFKGFFELFPSLSHNLLVD |
| 434 |
Q8TCX1 (DYNC2LI1) |
 Estimated TM-score=0.945 |
>Cytoplasmic dynein 2 light intermediate chain 1 (Length=351) MPSETLWEIAKAEVEKRGINGSEGDGAEIAEKFVFFIGSKNGGKTTIILRCLDRDEPPKP TLALEYTYGRRAKGHNTPKDIAHFWELGGGTSLLDLISIPITGDTLRTFSLVLVLDLSKP NDLWPTMENLLQATKSHVDKVIMKLGKTNAKAVSEMRQKIWNNMPKDHPDHELIDPFPVP LVIIGSKYDVFQDFESEKRKVICKTLRFVAHYYGASLMFTSKSEALLLKIRGVINQLAFG IDKSKSICVDQNKPLFITAGLDSFGQIGSPPVPENDIGKLHAHSPMELWKKVYEKLFPPK SINTLKDIKDPARDPQYAENEVDEMRIQKDLELEQYKRSSSKSWKQIELDS |
| 435 |
P20671 (H2AC7) |
 Estimated TM-score=0.945 |
>Histone H2A type 1-D (Length=130) MSGRGKQGGKARAKAKTRSSRAGLQFPVGRVHRLLRKGNYSERVGAGAPVYLAAVLEYLT AEILELAGNAARDNKKTRIIPRHLQLAIRNDEELNKLLGKVTIAQGGVLPNIQAVLLPKK TESHHKAKGK |
| 436 |
Q15274 (QPRT) |
 Estimated TM-score=0.945 |
>Nicotinate-nucleotide pyrophosphorylase [carboxylating] (Length=297) MDAEGLALLLPPVTLAALVDSWLREDCPGLNYAALVSGAGPSQAALWAKSPGVLAGQPFF DAIFTQLNCQVSWFLPEGSKLVPVARVAEVRGPAHCLLLGERVALNTLARCSGIASAAAA AVEAARGAGWTGHVAGTRKTTPGFRLVEKYGLLVGGAASHRYDLGGLVMVKDNHVVAAGG VEKAVRAARQAADFTLKVEVECSSLQEAVQAAEAGADLVLLDNFKPEELHPTATVLKAQF PSVAVEASGGITLDNLPQFCGPHIDVISMGMLTQAAPALDFSLKLFAKEVAPVPKIH |
| 437 |
Q9UM07 (PADI4) |
 Estimated TM-score=0.945 |
>Protein-arginine deiminase type-4 (Length=663) MAQGTLIRVTPEQPTHAVCVLGTLTQLDICSSAPEDCTSFSINASPGVVVDIAHGPPAKK KSTGSSTWPLDPGVEVTLTMKVASGSTGDQKVQISYYGPKTPPVKALLYLTGVEISLCAD ITRTGKVKPTRAVKDQRTWTWGPCGQGAILLVNCDRDNLESSAMDCEDDEVLDSEDLQDM SLMTLSTKTPKDFFTNHTLVLHVARSEMDKVRVFQATRGKLSSKCSVVLGPKWPSHYLMV PGGKHNMDFYVEALAFPDTDFPGLITLTISLLDTSNLELPEAVVFQDSVVFRVAPWIMTP NTQPPQEVYACSIFENEDFLKSVTTLAMKAKCKLTICPEEENMDDQWMQDEMEIGYIQAP HKTLPVVFDSPRNRGLKEFPIKRVMGPDFGYVTRGPQTGGISGLDSFGNLEVSPPVTVRG KEYPLGRILFGDSCYPSNDSRQMHQALQDFLSAQQVQAPVKLYSDWLSVGHVDEFLSFVP APDRKGFRLLLASPRSCYKLFQEQQNEGHGEALLFEGIKKKKQQKIKNILSNKTLREHNS FVERCIDWNRELLKRELGLAESDIIDIPQLFKLKEFSKAEAFFPNMVNMLVLGKHLGIPK PFGPVINGRCCLEEKVCSLLEPLGLQCTFINDFFTYHIRHGEVHCGTNVRRKPFSFKWWN MVP |
| 438 |
P13489 (RNH1) |
 Estimated TM-score=0.945 |
>Ribonuclease inhibitor (Length=461) MSLDIQSLDIQCEELSDARWAELLPLLQQCQVVRLDDCGLTEARCKDISSALRVNPALAE LNLRSNELGDVGVHCVLQGLQTPSCKIQKLSLQNCCLTGAGCGVLSSTLRTLPTLQELHL SDNLLGDAGLQLLCEGLLDPQCRLEKLQLEYCSLSAASCEPLASVLRAKPDFKELTVSNN DINEAGVRVLCQGLKDSPCQLEALKLESCGVTSDNCRDLCGIVASKASLRELALGSNKLG DVGMAELCPGLLHPSSRLRTLWIWECGITAKGCGDLCRVLRAKESLKELSLAGNELGDEG ARLLCETLLEPGCQLESLWVKSCSFTAACCSHFSSVLAQNRFLLELQISNNRLEDAGVRE LCQGLGQPGSVLRVLWLADCDVSDSSCSSLAATLLANHSLRELDLSNNCLGDAGILQLVE SVRQPGCLLEQLVLYDIYWSEEMEDRLQALEKDKPSLRVIS |
| 439 |
P04746 (AMY2A) |
 Estimated TM-score=0.944 |
>Pancreatic alpha-amylase (Length=511) MKFFLLLFTIGFCWAQYSPNTQQGRTSIVHLFEWRWVDIALECERYLAPKGFGGVQVSPP NENVAIYNPFRPWWERYQPVSYKLCTRSGNEDEFRNMVTRCNNVGVRIYVDAVINHMCGN AVSAGTSSTCGSYFNPGSRDFPAVPYSGWDFNDGKCKTGSGDIENYNDATQVRDCRLTGL LDLALEKDYVRSKIAEYMNHLIDIGVAGFRLDASKHMWPGDIKAILDKLHNLNSNWFPAG SKPFIYQEVIDLGGEPIKSSDYFGNGRVTEFKYGAKLGTVIRKWNGEKMSYLKNWGEGWG FVPSDRALVFVDNHDNQRGHGAGGASILTFWDARLYKMAVGFMLAHPYGFTRVMSSYRWP RQFQNGNDVNDWVGPPNNNGVIKEVTINPDTTCGNDWVCEHRWRQIRNMVIFRNVVDGQP FTNWYDNGSNQVAFGRGNRGFIVFNNDDWSFSLTLQTGLPAGTYCDVISGDKINGNCTGI KIYVSDDGKAHFSISNSAEDPFIAIHAESKL |
| 440 |
P61204 (ARF3) |
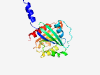 Estimated TM-score=0.944 |
>ADP-ribosylation factor 3 (Length=181) MGNIFGNLLKSLIGKKEMRILMVGLDAAGKTTILYKLKLGEIVTTIPTIGFNVETVEYKN ISFTVWDVGGQDKIRPLWRHYFQNTQGLIFVVDSNDRERVNEAREELMRMLAEDELRDAV LLVFANKQDLPNAMNAAEITDKLGLHSLRHRNWYIQATCATSGDGLYEGLDWLANQLKNK K |
| 441 |
P08133 (ANXA6) |
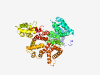 Estimated TM-score=0.944 |
>Annexin A6 (Length=673) MAKPAQGAKYRGSIHDFPGFDPNQDAEALYTAMKGFGSDKEAILDIITSRSNRQRQEVCQ SYKSLYGKDLIADLKYELTGKFERLIVGLMRPPAYCDAKEIKDAISGIGTDEKCLIEILA SRTNEQMHQLVAAYKDAYERDLEADIIGDTSGHFQKMLVVLLQGTREEDDVVSEDLVQQD VQDLYEAGELKWGTDEAQFIYILGNRSKQHLRLVFDEYLKTTGKPIEASIRGELSGDFEK LMLAVVKCIRSTPEYFAERLFKAMKGLGTRDNTLIRIMVSRSELDMLDIREIFRTKYEKS LYSMIKNDTSGEYKKTLLKLSGGDDDAAGQFFPEAAQVAYQMWELSAVARVELKGTVRPA NDFNPDADAKALRKAMKGLGTDEDTIIDIITHRSNVQRQQIRQTFKSHFGRDLMTDLKSE ISGDLARLILGLMMPPAHYDAKQLKKAMEGAGTDEKALIEILATRTNAEIRAINEAYKED YHKSLEDALSSDTSGHFRRILISLATGHREEGGENLDQAREDAQVAAEILEIADTPSGDK TSLETRFMTILCTRSYPHLRRVFQEFIKMTNYDVEHTIKKEMSGDVRDAFVAIVQSVKNK PLFFADKLYKSMKGAGTDEKTLTRIMVSRSEIDLLNIRREFIEKYDKSLHQAIEGDTSGD FLKALLALCGGED |
| 442 |
Q9UKL6 (PCTP) |
 Estimated TM-score=0.944 |
>Phosphatidylcholine transfer protein (Length=214) MELAAGSFSEEQFWEACAELQQPALAGADWQLLVETSGISIYRLLDKKTGLYEYKVFGVL EDCSPTLLADIYMDSDYRKQWDQYVKELYEQECNGETVVYWEVKYPFPMSNRDYVYLRQR RDLDMEGRKIHVILARSTSMPQLGERSGVIRVKQYKQSLAIESDGKKGSKVFMYYFDNPG GQIPSWLINWAAKNGVPNFLKDMARACQNYLKKT |
| 443 |
P06870 (KLK1) |
 Estimated TM-score=0.944 |
>Kallikrein-1 (Length=262) MWFLVLCLALSLGGTGAAPPIQSRIVGGWECEQHSQPWQAALYHFSTFQCGGILVHRQWV LTAAHCISDNYQLWLGRHNLFDDENTAQFVHVSESFPHPGFNMSLLENHTRQADEDYSHD LMLLRLTEPADTITDAVKVVELPTEEPEVGSTCLASGWGSIEPENFSFPDDLQCVDLKIL PNDECKKAHVQKVTDFMLCVGHLEGGKDTCVGDSGGPLMCDGVLQGVTSWGYVPCGTPNK PSVAVRVLSYVKWIEDTIAENS |
| 444 |
O75608 (LYPLA1) |
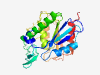 Estimated TM-score=0.944 |
>Acyl-protein thioesterase 1 (Length=230) MCGNNMSTPLPAIVPAARKATAAVIFLHGLGDTGHGWAEAFAGIRSSHIKYICPHAPVRP VTLNMNVAMPSWFDIIGLSPDSQEDESGIKQAAENIKALIDQEVKNGIPSNRIILGGFSQ GGALSLYTALTTQQKLAGVTALSCWLPLRASFPQGPIGGANRDISILQCHGDCDPLVPLM FGSLTVEKLKTLVNPANVTFKTYEGMMHSSCQQEMMDVKQFIDKLLPPID |
| 445 |
P47989 (XDH) |
 Estimated TM-score=0.944 |
>Xanthine dehydrogenase/oxidase (Length=1333) MTADKLVFFVNGRKVVEKNADPETTLLAYLRRKLGLSGTKLGCGEGGCGACTVMLSKYDR LQNKIVHFSANACLAPICSLHHVAVTTVEGIGSTKTRLHPVQERIAKSHGSQCGFCTPGI VMSMYTLLRNQPEPTMEEIENAFQGNLCRCTGYRPILQGFRTFARDGGCCGGDGNNPNCC MNQKKDHSVSLSPSLFKPEEFTPLDPTQEPIFPPELLRLKDTPRKQLRFEGERVTWIQAS TLKELLDLKAQHPDAKLVVGNTEIGIEMKFKNMLFPMIVCPAWIPELNSVEHGPDGISFG AACPLSIVEKTLVDAVAKLPAQKTEVFRGVLEQLRWFAGKQVKSVASVGGNIITASPISD LNPVFMASGAKLTLVSRGTRRTVQMDHTFFPGYRKTLLSPEEILLSIEIPYSREGEYFSA FKQASRREDDIAKVTSGMRVLFKPGTTEVQELALCYGGMANRTISALKTTQRQLSKLWKE ELLQDVCAGLAEELHLPPDAPGGMVDFRCTLTLSFFFKFYLTVLQKLGQENLEDKCGKLD PTFASATLLFQKDPPADVQLFQEVPKGQSEEDMVGRPLPHLAADMQASGEAVYCDDIPRY ENELSLRLVTSTRAHAKIKSIDTSEAKKVPGFVCFISADDVPGSNITGICNDETVFAKDK VTCVGHIIGAVVADTPEHTQRAAQGVKITYEELPAIITIEDAIKNNSFYGPELKIEKGDL KKGFSEADNVVSGEIYIGGQEHFYLETHCTIAVPKGEAGEMELFVSTQNTMKTQSFVAKM LGVPANRIVVRVKRMGGGFGGKETRSTVVSTAVALAAYKTGRPVRCMLDRDEDMLITGGR HPFLARYKVGFMKTGTVVALEVDHFSNVGNTQDLSQSIMERALFHMDNCYKIPNIRGTGR LCKTNLPSNTAFRGFGGPQGMLIAECWMSEVAVTCGMPAEEVRRKNLYKEGDLTHFNQKL EGFTLPRCWEECLASSQYHARKSEVDKFNKENCWKKRGLCIIPTKFGISFTVPFLNQAGA LLHVYTDGSVLLTHGGTEMGQGLHTKMVQVASRALKIPTSKIYISETSTNTVPNTSPTAA SVSADLNGQAVYAACQTILKRLEPYKKKNPSGSWEDWVTAAYMDTVSLSATGFYRTPNLG YSFETNSGNPFHYFSYGVACSEVEIDCLTGDHKNLRTDIVMDVGSSLNPAIDIGQVEGAF VQGLGLFTLEELHYSPEGSLHTRGPSTYKIPAFGSIPIEFRVSLLRDCPNKKAIYASKAV GEPPLFLAASIFFAIKDAIRAARAQHTGNNVKELFRLDSPATPEKIRNACVDKFTTLCVT GVPENCKPWSVRV |
| 446 |
P18124 (RPL7) |
 Estimated TM-score=0.944 |
>60S ribosomal protein L7 (Length=248) MEGVEEKKKEVPAVPETLKKKRRNFAELKIKRLRKKFAQKMLRKARRKLIYEKAKHYHKE YRQMYRTEIRMARMARKAGNFYVPAEPKLAFVIRIRGINGVSPKVRKVLQLLRLRQIFNG TFVKLNKASINMLRIVEPYIAWGYPNLKSVNELIYKRGYGKINKKRIALTDNALIARSLG KYGIICMEDLIHEIYTVGKRFKEANNFLWPFKLSSPRGGMKKKTTHFVEGGDAGNREDQI NRLIRRMN |
| 447 |
P54802 (NAGLU) |
 Estimated TM-score=0.944 |
>Alpha-N-acetylglucosaminidase (Length=743) MEAVAVAAAVGVLLLAGAGGAAGDEAREAAAVRALVARLLGPGPAADFSVSVERALAAKP GLDTYSLGGGGAARVRVRGSTGVAAAAGLHRYLRDFCGCHVAWSGSQLRLPRPLPAVPGE LTEATPNRYRYYQNVCTQSYSFVWWDWARWEREIDWMALNGINLALAWSGQEAIWQRVYL ALGLTQAEINEFFTGPAFLAWGRMGNLHTWDGPLPPSWHIKQLYLQHRVLDQMRSFGMTP VLPAFAGHVPEAVTRVFPQVNVTKMGSWGHFNCSYSCSFLLAPEDPIFPIIGSLFLRELI KEFGTDHIYGADTFNEMQPPSSEPSYLAAATTAVYEAMTAVDTEAVWLLQGWLFQHQPQF WGPAQIRAVLGAVPRGRLLVLDLFAESQPVYTRTASFQGQPFIWCMLHNFGGNHGLFGAL EAVNGGPEAARLFPNSTMVGTGMAPEGISQNEVVYSLMAELGWRKDPVPDLAAWVTSFAA RRYGVSHPDAGAAWRLLLRSVYNCSGEACRGHNRSPLVRRPSLQMNTSIWYNRSDVFEAW RLLLTSAPSLATSPAFRYDLLDLTRQAVQELVSLYYEEARSAYLSKELASLLRAGGVLAY ELLPALDEVLASDSRFLLGSWLEQARAAAVSEAEADFYEQNSRYQLTLWGPEGNILDYAN KQLAGLVANYYTPRWRLFLEALVDSVAQGIPFQQHQFDKNVFQLEQAFVLSKQRYPSQPR GDTVDLAKKIFLKYYPRWVAGSW |
| 448 |
P48637 (GSS) |
 Estimated TM-score=0.944 |
>Glutathione synthetase (Length=474) MATNWGSLLQDKQQLEELARQAVDRALAEGVLLRTSQEPTSSEVVSYAPFTLFPSLVPSA LLEQAYAVQMDFNLLVDAVSQNAAFLEQTLSSTIKQDDFTARLFDIHKQVLKEGIAQTVF LGLNRSDYMFQRSADGSPALKQIEINTISASFGGLASRTPAVHRHVLSVLSKTKEAGKIL SNNPSKGLALGIAKAWELYGSPNALVLLIAQEKERNIFDQRAIENELLARNIHVIRRTFE DISEKGSLDQDRRLFVDGQEIAVVYFRDGYMPRQYSLQNWEARLLLERSHAAKCPDIATQ LAGTKKVQQELSRPGMLEMLLPGQPEAVARLRATFAGLYSLDVGEEGDQAIAEALAAPSR FVLKPQREGGGNNLYGEEMVQALKQLKDSEERASYILMEKIEPEPFENCLLRPGSPARVV QCISELGIFGVYVRQEKTLVMNKHVGHLLRTKAIEHADGGVAAGVAVLDNPYPV |
| 449 |
P84077 (ARF1) |
 Estimated TM-score=0.943 |
>ADP-ribosylation factor 1 (Length=181) MGNIFANLFKGLFGKKEMRILMVGLDAAGKTTILYKLKLGEIVTTIPTIGFNVETVEYKN ISFTVWDVGGQDKIRPLWRHYFQNTQGLIFVVDSNDRERVNEAREELMRMLAEDELRDAV LLVFANKQDLPNAMNAAEITDKLGLHSLRHRNWYIQATCATSGDGLYEGLDWLSNQLRNQ K |
| 450 |
Q08209 (PPP3CA) |
 Estimated TM-score=0.943 |
>Serine/threonine-protein phosphatase 2B catalytic subunit alpha isoform (Length=521) MSEPKAIDPKLSTTDRVVKAVPFPPSHRLTAKEVFDNDGKPRVDILKAHLMKEGRLEESV ALRIITEGASILRQEKNLLDIDAPVTVCGDIHGQFFDLMKLFEVGGSPANTRYLFLGDYV DRGYFSIECVLYLWALKILYPKTLFLLRGNHECRHLTEYFTFKQECKIKYSERVYDACMD AFDCLPLAALMNQQFLCVHGGLSPEINTLDDIRKLDRFKEPPAYGPMCDILWSDPLEDFG NEKTQEHFTHNTVRGCSYFYSYPAVCEFLQHNNLLSILRAHEAQDAGYRMYRKSQTTGFP SLITIFSAPNYLDVYNNKAAVLKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEK VTEMLVNVLNICSDDELGSEEDGFDGATAAARKEVIRNKIRAIGKMARVFSVLREESESV LTLKGLTPTGMLPSGVLSGGKQTLQSATVEAIEADEAIKGFSPQHKITSFEEAKGLDRIN ERMPPRRDAMPSDANLNSINKALTSETNGTDSNGSNSSNIQ |
| 451 |
Q9BUP3 (HTATIP2) |
 Estimated TM-score=0.943 |
>Oxidoreductase HTATIP2 (Length=242) MAETEALSKLREDFRMQNKSVFILGASGETGRVLLKEILEQGLFSKVTLIGRRKLTFDEE AYKNVNQEVVDFEKLDDYASAFQGHDVGFCCLGTTRGKAGAEGFVRVDRDYVLKSAELAK AGGCKHFNLLSSKGADKSSNFLYLQVKGEVEAKVEELKFDRYSVFRPGVLLCDRQESRPG EWLVRKFFGSLPDSWASGHSVPVVTVVRAMLNNVVRPRDKQMELLENKAIHDLGKAHGSL KP |
| 452 |
P33552 (CKS2) |
 Estimated TM-score=0.943 |
>Cyclin-dependent kinases regulatory subunit 2 (Length=79) MAHKQIYYSDKYFDEHYEYRHVMLPRELSKQVPKTHLMSEEEWRRLGVQQSLGWVHYMIH EPEPHILLFRRPLPKDQQK |
| 453 |
P49720 (PSMB3) |
 Estimated TM-score=0.943 |
>Proteasome subunit beta type-3 (Length=205) MSIMSYNGGAVMAMKGKNCVAIAADRRFGIQAQMVTTDFQKIFPMGDRLYIGLAGLATDV QTVAQRLKFRLNLYELKEGRQIKPYTLMSMVANLLYEKRFGPYYTEPVIAGLDPKTFKPF ICSLDLIGCPMVTDDFVVSGTCAEQMYGMCESLWEPNMDPDHLFETISQAMLNAVDRDAV SGMGVIVHIIEKDKITTRTLKARMD |
| 454 |
P62745 (RHOB) |
 Estimated TM-score=0.943 |
>Rho-related GTP-binding protein RhoB (Length=196) MAAIRKKLVVVGDGACGKTCLLIVFSKDEFPEVYVPTVFENYVADIEVDGKQVELALWDT AGQEDYDRLRPLSYPDTDVILMCFSVDSPDSLENIPEKWVPEVKHFCPNVPIILVANKKD LRSDEHVRTELARMKQEPVRTDDGRAMAVRIQAYDYLECSAKTKEGVREVFETATRAALQ KRYGSQNGCINCCKVL |
| 455 |
P30556 (AGTR1) |
 Estimated TM-score=0.943 |
>Type-1 angiotensin II receptor (Length=359) MILNSSTEDGIKRIQDDCPKAGRHNYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLK TVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLT CLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHRNVFFIENTNITVC AFHYESQNSTLPIGLGLTKNILGFLFPFLIILTSYTLIWKALKKAYEIQKNKPRNDDIFK IIMAIVLFFFFSWIPHQIFTFLDVLIQLGIIRDCRIADIVDTAMPITICIAYFNNCLNPL FYGFLGKKFKRYFLQLLKYIPPKAKSHSNLSTKMSTLSYRPSDNVSSSTKKPAPCFEVE |
| 456 |
Q9NZL9 (MAT2B) |
 Estimated TM-score=0.943 |
>Methionine adenosyltransferase 2 subunit beta (Length=334) MVGREKELSIHFVPGSCRLVEEEVNIPNRRVLVTGATGLLGRAVHKEFQQNNWHAVGCGF RRARPKFEQVNLLDSNAVHHIIHDFQPHVIVHCAAERRPDVVENQPDAASQLNVDASGNL AKEAAAVGAFLIYISSDYVFDGTNPPYREEDIPAPLNLYGKTKLDGEKAVLENNLGAAVL RIPILYGEVEKLEESAVTVMFDKVQFSNKSANMDHWQQRFPTHVKDVATVCRQLAEKRML DPSIKGTFHWSGNEQMTKYEMACAIADAFNLPSSHLRPITDSPVLGAQRPRNAQLDCSKL ETLGIGQRTPFRIGIKESLWPFLIDKRWRQTVFH |
| 457 |
P36404 (ARL2) |
 Estimated TM-score=0.943 |
>ADP-ribosylation factor-like protein 2 (Length=184) MGLLTILKKMKQKERELRLLMLGLDNAGKTTILKKFNGEDIDTISPTLGFNIKTLEHRGF KLNIWDVGGQKSLRSYWRNYFESTDGLIWVVDSADRQRMQDCQRELQSLLVEERLAGATL LIFANKQDLPGALSSNAIREVLELDSIRSHHWCIQGCSAVTGENLLPGIDWLLDDISSRI FTAD |
| 458 |
O95989 (NUDT3) |
 Estimated TM-score=0.943 |
>Diphosphoinositol polyphosphate phosphohydrolase 1 (Length=172) MMKLKSNQTRTYDGDGYKKRAACLCFRSESEEEVLLVSSSRHPDRWIVPGGGMEPEEEPS VAAVREVCEEAGVKGTLGRLVGIFENQERKHRTYVYVLIVTEVLEDWEDSVNIGRKREWF KIEDAIKVLQYHKPVQASYFETLRQGYSANNGTPVVATTYSVSAQSSMSGIR |
| 459 |
P07288 (KLK3) |
 Estimated TM-score=0.943 |
>Prostate-specific antigen (Length=261) MWVPVVFLTLSVTWIGAAPLILSRIVGGWECEKHSQPWQVLVASRGRAVCGGVLVHPQWV LTAAHCIRNKSVILLGRHSLFHPEDTGQVFQVSHSFPHPLYDMSLLKNRFLRPGDDSSHD LMLLRLSEPAELTDAVKVMDLPTQEPALGTTCYASGWGSIEPEEFLTPKKLQCVDLHVIS NDVCAQVHPQKVTKFMLCAGRWTGGKSTCSGDSGGPLVCNGVLQGITSWGSEPCALPERP SLYTKVVHYRKWIKDTIVANP |
| 460 |
P07311 (ACYP1) |
 Estimated TM-score=0.943 |
>Acylphosphatase-1 (Length=99) MAEGNTLISVDYEIFGKVQGVFFRKHTQAEGKKLGLVGWVQNTDRGTVQGQLQGPISKVR HMQEWLETRGSPKSHIDKANFNNEKVILKLDYSDFQIVK |
| 461 |
P30038 (ALDH4A1) |
 Estimated TM-score=0.943 |
>Delta-1-pyrroline-5-carboxylate dehydrogenase, mitochondrial (Length=563) MLLPAPALRRALLSRPWTGAGLRWKHTSSLKVANEPVLAFTQGSPERDALQKALKDLKGR MEAIPCVVGDEEVWTSDVQYQVSPFNHGHKVAKFCYADKSLLNKAIEAALAARKEWDLKP IADRAQIFLKAADMLSGPRRAEILAKTMVGQGKTVIQAEIDAAAELIDFFRFNAKYAVEL EGQQPISVPPSTNSTVYRGLEGFVAAISPFNFTAIGGNLAGAPALMGNVVLWKPSDTAML ASYAVYRILREAGLPPNIIQFVPADGPLFGDTVTSSEHLCGINFTGSVPTFKHLWKQVAQ NLDRFHTFPRLAGECGGKNFHFVHRSADVESVVSGTLRSAFEYGGQKCSACSRLYVPHSL WPQIKGRLLEEHSRIKVGDPAEDFGTFFSAVIDAKSFARIKKWLEHARSSPSLTILAGGK CDDSVGYFVEPCIVESKDPQEPIMKEEIFGPVLSVYVYPDDKYKETLQLVDSTTSYGLTG AVFSQDKDVVQEATKVLRNAAGNFYINDKSTGSIVGQQPFGGARASGTNDKPGGPHYILR WTSPQVIKETHKPLGDWSYAYMQ |
| 462 |
Q13895 (BYSL) |
 Estimated TM-score=0.943 |
>Bystin (Length=437) MPKFKAARGVGGQEKHAPLADQILAGNAVRAGVREKRRGRGTGEAEEEYVGPRLSRRILQ QARQQQEELEAEHGTGDKPAAPRERTTRLGPRMPQDGSDDEDEEWPTLEKAATMTAAGHH AEVVVDPEDERAIEMFMNKNPPARRTLADIIMEKLTEKQTEVETVMSEVSGFPMPQLDPR VLEVYRGVREVLSKYRSGKLPKAFKIIPALSNWEQILYVTEPEAWTAAAMYQATRIFASN LKERMAQRFYNLVLLPRVRDDVAEYKRLNFHLYMALKKALFKPGAWFKGILIPLCESGTC TLREAIIVGSIITKCSIPVLHSSAAMLKIAEMEYSGANSIFLRLLLDKKYALPYRVLDAL VFHFLGFRTEKRELPVLWHQCLLTLVQRYKADLATDQKEALLELLRLQPHPQLSPEIRRE LQSAVPRDVEDVPITVE |
| 463 |
Q96F10 (SAT2) |
 Estimated TM-score=0.943 |
>Diamine acetyltransferase 2 (Length=170) MASVRIREAKEGDCGDILRLIRELAEFEKLSDQVKISEEALRADGFGDNPFYHCLVAEIL PAPGKLLGPCVVGYGIYYFIYSTWKGRTIYLEDIYVMPEYRGQGIGSKIIKKVAEVALDK GCSQFRLAVLDWNQRAMDLYKALGAQDLTEAEGWHFFCFQGEATRKLAGK |
| 464 |
Q08257 (CRYZ) |
 Estimated TM-score=0.943 |
>Quinone oxidoreductase (Length=329) MATGQKLMRAVRVFEFGGPEVLKLRSDIAVPIPKDHQVLIKVHACGVNPVETYIRSGTYS RKPLLPYTPGSDVAGVIEAVGDNASAFKKGDRVFTSSTISGGYAEYALAADHTVYKLPEK LDFKQGAAIGIPYFTAYRALIHSACVKAGESVLVHGASGGVGLAACQIARAYGLKILGTA GTEEGQKIVLQNGAHEVFNHREVNYIDKIKKYVGEKGIDIIIEMLANVNLSKDLSLLSHG GRVIVVGSRGTIEINPRDTMAKESSIIGVTLFSSTKEEFQQYAAALQAGMEIGWLKPVIG SQYPLEKVAEAHENIIHGSGATGKMILLL |
| 465 |
Q9BSH5 (HDHD3) |
 Estimated TM-score=0.943 |
>Haloacid dehalogenase-like hydrolase domain-containing protein 3 (Length=251) MAHRLQIRLLTWDVKDTLLRLRHPLGEAYATKARAHGLEVEPSALEQGFRQAYRAQSHSF PNYGLSHGLTSRQWWLDVVLQTFHLAGVQDAQAVAPIAEQLYKDFSHPCTWQVLDGAEDT LRECRTRGLRLAVISNFDRRLEGILGGLGLREHFDFVLTSEAAGWPKPDPRIFQEALRLA HMEPVVAAHVGDNYLCDYQGPRAVGMHSFLVVGPQALDPVVRDSVPKEHILPSLAHLLPA LDCLEGSTPGL |
| 466 |
Q15771 (RAB30) |
 Estimated TM-score=0.943 |
>Ras-related protein Rab-30 (Length=203) MSMEDYDFLFKIVLIGNAGVGKTCLVRRFTQGLFPPGQGATIGVDFMIKTVEINGEKVKL QIWDTAGQERFRSITQSYYRSANALILTYDITCEESFRCLPEWLREIEQYASNKVITVLV GNKIDLAERREVSQQRAEEFSEAQDMYYLETSAKESDNVEKLFLDLACRLISEARQNTLV NNVSSPLPGEGKSISYLTCCNFN |
| 467 |
P08758 (ANXA5) |
 Estimated TM-score=0.943 |
>Annexin A5 (Length=320) MAQVLRGTVTDFPGFDERADAETLRKAMKGLGTDEESILTLLTSRSNAQRQEISAAFKTL FGRDLLDDLKSELTGKFEKLIVALMKPSRLYDAYELKHALKGAGTNEKVLTEIIASRTPE ELRAIKQVYEEEYGSSLEDDVVGDTSGYYQRMLVVLLQANRDPDAGIDEAQVEQDAQALF QAGELKWGTDEEKFITIFGTRSVSHLRKVFDKYMTISGFQIEETIDRETSGNLEQLLLAV VKSIRSIPAYLAETLYYAMKGAGTDDHTLIRVMVSRSEIDLFNIRKEFRKNFATSLYSMI KGDTSGDYKKALLLLCGEDD |
| 468 |
P02792 (FTL) |
 Estimated TM-score=0.942 |
>Ferritin light chain (Length=175) MSSQIRQNYSTDVEAAVNSLVNLYLQASYTYLSLGFYFDRDDVALEGVSHFFRELAEEKR EGYERLLKMQNQRGGRALFQDIKKPAEDEWGKTPDAMKAAMALEKKLNQALLDLHALGSA RTDPHLCDFLETHFLDEEVKLIKKMGDHLTNLHRLGGPEAGLGEYLFERLTLKHD |
| 469 |
Q9NRG7 (SDR39U1) |
 Estimated TM-score=0.942 |
>Epimerase family protein SDR39U1 (Length=293) MRVLVGGGTGFIGTALTQLLNARGHEVTLVSRKPGPGRITWDELAASGLPSCDAAVNLAG ENILNPLRRWNETFQKEVIGSRLETTQLLAKAITKAPQPPKAWVLVTGVAYYQPSLTAEY DEDSPGGDFDFFSNLVTKWEAAARLPGDSTRQVVVRSGVVLGRGGGAMGHMLLPFRLGLG GPIGSGHQFFPWIHIGDLAGILTHALEANHVHGVLNGVAPSSATNAEFAQTLGAALGRRA FIPLPSAVVQAVFGRQRAIMLLEGQKVIPQRTLATGYQYSFPELGAALKEIVA |
| 470 |
P62834 (RAP1A) |
 Estimated TM-score=0.942 |
>Ras-related protein Rap-1A (Length=184) MREYKLVVLGSGGVGKSALTVQFVQGIFVEKYDPTIEDSYRKQVEVDCQQCMLEILDTAG TEQFTAMRDLYMKNGQGFALVYSITAQSTFNDLQDLREQILRVKDTEDVPMILVGNKCDL EDERVVGKEQGQNLARQWCNCAFLESSAKSKINVNEIFYDLVRQINRKTPVEKKKPKKKS CLLL |
| 471 |
P60953 (CDC42) |
 Estimated TM-score=0.942 |
>Cell division control protein 42 homolog (Length=191) MQTIKCVVVGDGAVGKTCLLISYTTNKFPSEYVPTVFDNYAVTVMIGGEPYTLGLFDTAG QEDYDRLRPLSYPQTDVFLVCFSVVSPSSFENVKEKWVPEITHHCPKTPFLLVGTQIDLR DDPSTIEKLAKNKQKPITPETAEKLARDLKAVKYVECSALTQKGLKNVFDEAILAALEPP EPKKSRRCVLL |
| 472 |
P62942 (FKBP1A) |
 Estimated TM-score=0.942 |
>Peptidyl-prolyl cis-trans isomerase FKBP1A (Length=108) MGVQVETISPGDGRTFPKRGQTCVVHYTGMLEDGKKFDSSRDRNKPFKFMLGKQEVIRGW EEGVAQMSVGQRAKLTISPDYAYGATGHPGIIPPHATLVFDVELLKLE |
| 473 |
P51648 (ALDH3A2) |
 Estimated TM-score=0.942 |
>Aldehyde dehydrogenase family 3 member A2 (Length=485) MELEVRRVRQAFLSGRSRPLRFRLQQLEALRRMVQEREKDILTAIAADLCKSEFNVYSQE VITVLGEIDFMLENLPEWVTAKPVKKNVLTMLDEAYIQPQPLGVVLIIGAWNYPFVLTIQ PLIGAIAAGNAVIIKPSELSENTAKILAKLLPQYLDQDLYIVINGGVEETTELLKQRFDH IFYTGNTAVGKIVMEAAAKHLTPVTLELGGKSPCYIDKDCDLDIVCRRITWGKYMNCGQT CIAPDYILCEASLQNQIVWKIKETVKEFYGENIKESPDYERIINLRHFKRILSLLEGQKI AFGGETDEATRYIAPTVLTDVDPKTKVMQEEIFGPILPIVPVKNVDEAINFINEREKPLA LYVFSHNHKLIKRMIDETSSGGVTGNDVIMHFTLNSFPFGGVGSSGMGAYHGKHSFDTFS HQRPCLLKSLKREGANKLRYPPNSQSKVDWGKFFLLKRFNKEKLGLLLLTFLGIVAAVLV KAEYY |
| 474 |
P05181 (CYP2E1) |
 Estimated TM-score=0.942 |
>Cytochrome P450 2E1 (Length=493) MSALGVTVALLVWAAFLLLVSMWRQVHSSWNLPPGPFPLPIIGNLFQLELKNIPKSFTRL AQRFGPVFTLYVGSQRMVVMHGYKAVKEALLDYKDEFSGRGDLPAFHAHRDRGIIFNNGP TWKDIRRFSLTTLRNYGMGKQGNESRIQREAHFLLEALRKTQGQPFDPTFLIGCAPCNVI ADILFRKHFDYNDEKFLRLMYLFNENFHLLSTPWLQLYNNFPSFLHYLPGSHRKVIKNVA EVKEYVSERVKEHHQSLDPNCPRDLTDCLLVEMEKEKHSAERLYTMDGITVTVADLFFAG TETTSTTLRYGLLILMKYPEIEEKLHEEIDRVIGPSRIPAIKDRQEMPYMDAVVHEIQRF ITLVPSNLPHEATRDTIFRGYLIPKGTVVVPTLDSVLYDNQEFPDPEKFKPEHFLNENGK FKYSDYFKPFSTGKRVCAGEGLARMELFLLLCAILQHFNLKPLVDPKDIDLSPIHIGFGC IPPRYKLCVIPRS |
| 475 |
Q9H7B4 (SMYD3) |
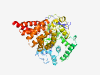 Estimated TM-score=0.942 |
>Histone-lysine N-methyltransferase SMYD3 (Length=428) MEPLKVEKFATAKRGNGLRAVTPLRPGELLFRSDPLAYTVCKGSRGVVCDRCLLGKEKLM RCSQCRVAKYCSAKCQKKAWPDHKRECKCLKSCKPRYPPDSVRLLGRVVFKLMDGAPSES EKLYSFYDLESNINKLTEDKKEGLRQLVMTFQHFMREEIQDASQLPPAFDLFEAFAKVIC NSFTICNAEMQEVGVGLYPSISLLNHSCDPNCSIVFNGPHLLLRAVRDIEVGEELTICYL DMLMTSEERRKQLRDQYCFECDCFRCQTQDKDADMLTGDEQVWKEVQESLKKIEELKAHW KWEQVLAMCQAIISSNSERLPDINIYQLKVLDCAMDACINLGLLEEALFYGTRTMEPYRI FFPGSHPVRGVQVMKVGKLQLHQGMFPQAMKNLRLAFDIMRVTHGREHSLIEDLILLLEE CDANIRAS |
| 476 |
P18621 (RPL17) |
 Estimated TM-score=0.942 |
>60S ribosomal protein L17 (Length=184) MVRYSLDPENPTKSCKSRGSNLRVHFKNTRETAQAIKGMHIRKATKYLKDVTLQKQCVPF RRYNGGVGRCAQAKQWGWTQGRWPKKSAEFLLHMLKNAESNAELKGLDVDSLVIEHIQVN KAPKMRRRTYRAHGRINPYMSSPCHIEMILTEKEQIVPKPEEEVAQKKKISQKKLKKQKL MARE |
| 477 |
Q9UBQ0 (VPS29) |
 Estimated TM-score=0.942 |
>Vacuolar protein sorting-associated protein 29 (Length=182) MLVLVLGDLHIPHRCNSLPAKFKKLLVPGKIQHILCTGNLCTKESYDYLKTLAGDVHIVR GDFDENLNYPEQKVVTVGQFKIGLIHGHQVIPWGDMASLALLQRQFDVDILISGHTHKFE AFEHENKFYINPGSATGAYNALETNIIPSFVLMDIQASTVVTYVYQLIGDDVKVERIEYK KP |
| 478 |
Q9UHA4 (LAMTOR3) |
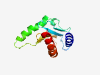 Estimated TM-score=0.942 |
>Ragulator complex protein LAMTOR3 (Length=124) MADDLKRFLYKKLPSVEGLHAIVVSDRDGVPVIKVANDNAPEHALRPGFLSTFALATDQG SKLGLSKNKSIICYYNTYQVVQFNRLPLVVSFIASSSANTGLIVSLEKELAPLFEELRQV VEVS |
| 479 |
P17050 (NAGA) |
 Estimated TM-score=0.942 |
>Alpha-N-acetylgalactosaminidase (Length=411) MLLKTVLLLGHVAQVLMLDNGLLQTPPMGWLAWERFRCNINCDEDPKNCISEQLFMEMAD RMAQDGWRDMGYTYLNIDDCWIGGRDASGRLMPDPKRFPHGIPFLADYVHSLGLKLGIYA DMGNFTCMGYPGTTLDKVVQDAQTFAEWKVDMLKLDGCFSTPEERAQGYPKMAAALNATG RPIAFSCSWPAYEGGLPPRVNYSLLADICNLWRNYDDIQDSWWSVLSILNWFVEHQDILQ PVAGPGHWNDPDMLLIGNFGLSLEQSRAQMALWTVLAAPLLMSTDLRTISAQNMDILQNP LMIKINQDPLGIQGRRIHKEKSLIEVYMRPLSNKASALVFFSCRTDMPYRYHSSLGQLNF TGSVIYEAQDVYSGDIISGLRDETNFTVIINPSGVVMWYLYPIKNLEMSQQ |
| 480 |
O75817 (POP7) |
 Estimated TM-score=0.942 |
>Ribonuclease P protein subunit p20 (Length=140) MAENREPRGAVEAELDPVEYTLRKRLPSRLPRRPNDIYVNMKTDFKAQLARCQKLLDGGA RGQNACSEIYIHGLGLAINRAINIALQLQAGSFGSLQVAANTSTVELVDELEPETDTREP LTRIRNNSAIHIRVFRVTPK |
| 481 |
P14618 (PKM) |
 Estimated TM-score=0.942 |
>Pyruvate kinase PKM (Length=531) MSKPHSEAGTAFIQTQQLHAAMADTFLEHMCRLDIDSPPITARNTGIICTIGPASRSVET LKEMIKSGMNVARLNFSHGTHEYHAETIKNVRTATESFASDPILYRPVAVALDTKGPEIR TGLIKGSGTAEVELKKGATLKITLDNAYMEKCDENILWLDYKNICKVVEVGSKIYVDDGL ISLQVKQKGADFLVTEVENGGSLGSKKGVNLPGAAVDLPAVSEKDIQDLKFGVEQDVDMV FASFIRKASDVHEVRKVLGEKGKNIKIISKIENHEGVRRFDEILEASDGIMVARGDLGIE IPAEKVFLAQKMMIGRCNRAGKPVICATQMLESMIKKPRPTRAEGSDVANAVLDGADCIM LSGETAKGDYPLEAVRMQHLIAREAEAAIYHLQLFEELRRLAPITSDPTEATAVGAVEAS FKCCSGAIIVLTKSGRSAHQVARYRPRAPIIAVTRNPQTARQAHLYRGIFPVLCKDPVQE AWAEDVDLRVNFAMNVGKARGFFKKGDVVIVLTGWRPGSGFTNTMRVVPVP |
| 482 |
O95045 (UPP2) |
 Estimated TM-score=0.942 |
>Uridine phosphorylase 2 (Length=317) MASVIPASNRSMRSDRNTYVGKRFVHVKNPYLDLMDEDILYHLDLGTKTHNLPAMFGDVK FVCVGGSPNRMKAFALFMHKELGFEEAEEDIKDICAGTDRYCMYKTGPVLAISHGMGIPS ISIMLHELIKLLHHARCCDVTIIRIGTSGGIGIAPGTVVITDIAVDSFFKPRFEQVILDN IVTRSTELDKELSEELFNCSKEIPNFPTLVGHTMCTYDFYEGQGRLDGALCSFSREKKLD YLKRAFKAGVRNIEMESTVFAAMCGLCGLKAAVVCVTLLDRLDCDQINLPHDVLVEYQQR PQLLISNFIRRRLGLCD |
| 483 |
Q07020 (RPL18) |
 Estimated TM-score=0.941 |
>60S ribosomal protein L18 (Length=188) MGVDIRHNKDRKVRRKEPKSQDIYLRLLVKLYRFLARRTNSTFNQVVLKRLFMSRTNRPP LSLSRMIRKMKLPGRENKTAVVVGTITDDVRVQEVPKLKVCALRVTSRARSRILRAGGKI LTFDQLALDSPKGCGTVLLSGPRKGREVYRHFGKAPGTPHSHTKPYVRSKGRKFERARGR RASRGYKN |
| 484 |
O60547 (GMDS) |
 Estimated TM-score=0.941 |
>GDP-mannose 4,6 dehydratase (Length=372) MAHAPARCPSARGSGDGEMGKPRNVALITGITGQDGSYLAEFLLEKGYEVHGIVRRSSSF NTGRIEHLYKNPQAHIEGNMKLHYGDLTDSTCLVKIINEVKPTEIYNLGAQSHVKISFDL AEYTADVDGVGTLRLLDAVKTCGLINSVKFYQASTSELYGKVQEIPQKETTPFYPRSPYG AAKLYAYWIVVNFREAYNLFAVNGILFNHESPRRGANFVTRKISRSVAKIYLGQLECFSL GNLDAKRDWGHAKDYVEAMWLMLQNDEPEDFVIATGEVHSVREFVEKSFLHIGKTIVWEG KNENEVGRCKETGKVHVTVDLKYYRPTEVDFLQGDCTKAKQKLNWKPRVAFDELVREMVH ADVELMRTNPNA |
| 485 |
Q9BV86 (NTMT1) |
 Estimated TM-score=0.941 |
>N-terminal Xaa-Pro-Lys N-methyltransferase 1 (Length=223) MTSEVIEDEKQFYSKAKTYWKQIPPTVDGMLGGYGHISSIDINSSRKFLQRFLREGPNKT GTSCALDCGAGIGRITKRLLLPLFREVDMVDITEDFLVQAKTYLGEEGKRVRNYFCCGLQ DFTPEPDSYDVIWIQWVIGHLTDQHLAEFLRRCKGSLRPNGIIVIKDNMAQEGVILDDVD SSVCRDLDVVRRIICSAGLSLLAEERQENLPDEIYHVYSFALR |
| 486 |
Q4G0W2 (DUSP28) |
 Estimated TM-score=0.941 |
>Dual specificity phosphatase 28 (Length=176) MGPAEAGRRGAASPVPPPLVRVAPSLFLGSARAAGAEEQLARAGVTLCVNVSRQQPGPRA PGVAELRVPVFDDPAEDLLAHLEPTCAAMEAAVRAGGACLVYCKNGRSRSAAVCTAYLMR HRGLSLAKAFQMVKSARPVAEPNPGFWSQLQKYEEALQAQSCLQGEPPALGLGPEA |
| 487 |
P60520 (GABARAPL2) |
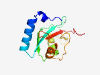 Estimated TM-score=0.941 |
>Gamma-aminobutyric acid receptor-associated protein-like 2 (Length=117) MKWMFKEDHSLEHRCVESAKIRAKYPDRVPVIVEKVSGSQIVDIDKRKYLVPSDITVAQF MWIIRKRIQLPSEKAIFLFVDKTVPQSSLTMGQLYEKEKDEDGFLYVAYSGENTFGF |
| 488 |
O75874 (IDH1) |
 Estimated TM-score=0.941 |
>Isocitrate dehydrogenase [NADP] cytoplasmic (Length=414) MSKKISGGSVVEMQGDEMTRIIWELIKEKLIFPYVELDLHSYDLGIENRDATNDQVTKDA AEAIKKHNVGVKCATITPDEKRVEEFKLKQMWKSPNGTIRNILGGTVFREAIICKNIPRL VSGWVKPIIIGRHAYGDQYRATDFVVPGPGKVEITYTPSDGTQKVTYLVHNFEEGGGVAM GMYNQDKSIEDFAHSSFQMALSKGWPLYLSTKNTILKKYDGRFKDIFQEIYDKQYKSQFE AQKIWYEHRLIDDMVAQAMKSEGGFIWACKNYDGDVQSDSVAQGYGSLGMMTSVLVCPDG KTVEAEAAHGTVTRHYRMYQKGQETSTNPIASIFAWTRGLAHRAKLDNNKELAFFANALE EVSIETIEAGFMTKDLAACIKGLPNVQRSDYLNTFEFMDKLGENLKIKLAQAKL |
| 489 |
P52209 (PGD) |
 Estimated TM-score=0.941 |
>6-phosphogluconate dehydrogenase, decarboxylating (Length=483) MAQADIALIGLAVMGQNLILNMNDHGFVVCAFNRTVSKVDDFLANEAKGTKVVGAQSLKE MVSKLKKPRRIILLVKAGQAVDDFIEKLVPLLDTGDIIIDGGNSEYRDTTRRCRDLKAKG ILFVGSGVSGGEEGARYGPSLMPGGNKEAWPHIKTIFQGIAAKVGTGEPCCDWVGDEGAG HFVKMVHNGIEYGDMQLICEAYHLMKDVLGMAQDEMAQAFEDWNKTELDSFLIEITANIL KFQDTDGKHLLPKIRDSAGQKGTGKWTAISALEYGVPVTLIGEAVFARCLSSLKDERIQA SKKLKGPQKFQFDGDKKSFLEDIRKALYASKIISYAQGFMLLRQAATEFGWTLNYGGIAL MWRGGCIIRSVFLGKIKDAFDRNPELQNLLLDDFFKSAVENCQDSWRRAVSTGVQAGIPM PCFTTALSFYDGYRHEMLPASLIQAQRDYFGAHTYELLAKPGQFIHTNWTGHGGTVSSSS YNA |
| 490 |
P62330 (ARF6) |
 Estimated TM-score=0.941 |
>ADP-ribosylation factor 6 (Length=175) MGKVLSKIFGNKEMRILMLGLDAAGKTTILYKLKLGQSVTTIPTVGFNVETVTYKNVKFN VWDVGGQDKIRPLWRHYYTGTQGLIFVVDCADRDRIDEARQELHRIINDREMRDAIILIF ANKQDLPDAMKPHEIQEKLGLTRIRDRNWYVQPSCATSGDGLYEGLTWLTSNYKS |
| 491 |
O95166 (GABARAP) |
 Estimated TM-score=0.941 |
>Gamma-aminobutyric acid receptor-associated protein (Length=117) MKFVYKEEHPFEKRRSEGEKIRKKYPDRVPVIVEKAPKARIGDLDKKKYLVPSDLTVGQF YFLIRKRIHLRAEDALFFFVNNVIPPTSATMGQLYQEHHEEDFFLYIAYSDESVYGL |
| 492 |
P09211 (GSTP1) |
 Estimated TM-score=0.941 |
>Glutathione S-transferase P (Length=210) MPPYTVVYFPVRGRCAALRMLLADQGQSWKEEVVTVETWQEGSLKASCLYGQLPKFQDGD LTLYQSNTILRHLGRTLGLYGKDQQEAALVDMVNDGVEDLRCKYISLIYTNYEAGKDDYV KALPGQLKPFETLLSQNQGGKTFIVGDQISFADYNLLDLLLIHEVLAPGCLDAFPLLSAY VGRLSARPKLKAFLASPEYVNLPINGNGKQ |
| 493 |
P05177 (CYP1A2) |
 Estimated TM-score=0.941 |
>Cytochrome P450 1A2 (Length=516) MALSQSVPFSATELLLASAIFCLVFWVLKGLRPRVPKGLKSPPEPWGWPLLGHVLTLGKN PHLALSRMSQRYGDVLQIRIGSTPVLVLSRLDTIRQALVRQGDDFKGRPDLYTSTLITDG QSLTFSTDSGPVWAARRRLAQNALNTFSIASDPASSSSCYLEEHVSKEAKALISRLQELM AGPGHFDPYNQVVVSVANVIGAMCFGQHFPESSDEMLSLVKNTHEFVETASSGNPLDFFP ILRYLPNPALQRFKAFNQRFLWFLQKTVQEHYQDFDKNSVRDITGALFKHSKKGPRASGN LIPQEKIVNLVNDIFGAGFDTVTTAISWSLMYLVTKPEIQRKIQKELDTVIGRERRPRLS DRPQLPYLEAFILETFRHSSFLPFTIPHSTTRDTTLNGFYIPKKCCVFVNQWQVNHDPEL WEDPSEFRPERFLTADGTAINKPLSEKMMLFGMGKRRCIGEVLAKWEIFLFLAILLQQLE FSVPPGVKVDLTPIYGLTMKHARCEHVQARLRFSIN |
| 494 |
Q00266 (MAT1A) |
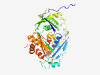 Estimated TM-score=0.941 |
>S-adenosylmethionine synthase isoform type-1 (Length=395) MNGPVDGLCDHSLSEGVFMFTSESVGEGHPDKICDQISDAVLDAHLKQDPNAKVACETVC KTGMVLLCGEITSMAMVDYQRVVRDTIKHIGYDDSAKGFDFKTCNVLVALEQQSPDIAQC VHLDRNEEDVGAGDQGLMFGYATDETEECMPLTIILAHKLNARMADLRRSGLLPWLRPDS KTQVTVQYMQDNGAVIPVRIHTIVISVQHNEDITLEEMRRALKEQVIRAVVPAKYLDEDT VYHLQPSGRFVIGGPQGDAGVTGRKIIVDTYGGWGAHGGGAFSGKDYTKVDRSAAYAARW VAKSLVKAGLCRRVLVQVSYAIGVAEPLSISIFTYGTSQKTERELLDVVHKNFDLRPGVI VRDLDLKKPIYQKTACYGHFGRSEFPWEVPRKLVF |
| 495 |
P20132 (SDS) |
 Estimated TM-score=0.940 |
>L-serine dehydratase/L-threonine deaminase (Length=328) MMSGEPLHVKTPIRDSMALSKMAGTSVYLKMDSAQPSGSFKIRGIGHFCKRWAKQGCAHF VCSSAGNAGMAAAYAARQLGVPATIVVPSTTPALTIERLKNEGATVKVVGELLDEAFELA KALAKNNPGWVYIPPFDDPLIWEGHASIVKELKETLWEKPGAIALSVGGGGLLCGVVQGL QEVGWGDVPVIAMETFGAHSFHAATTAGKLVSLPKITSVAKALGVKTVGAQALKLFQEHP IFSEVISDQEAVAAIEKFVDDEKILVEPACGAALAAVYSHVIQKLQLEGNLRTPLPSLVV IVCGGSNISLAQLRALKEQLGMTNRLPK |
| 496 |
P22680 (CYP7A1) |
 Estimated TM-score=0.940 |
>Cytochrome P450 7A1 (Length=504) MMTTSLIWGIAIAACCCLWLILGIRRRQTGEPPLENGLIPYLGCALQFGANPLEFLRANQ RKHGHVFTCKLMGKYVHFITNPLSYHKVLCHGKYFDWKKFHFATSAKAFGHRSIDPMDGN TTENINDTFIKTLQGHALNSLTESMMENLQRIMRPPVSSNSKTAAWVTEGMYSFCYRVMF EAGYLTIFGRDLTRRDTQKAHILNNLDNFKQFDKVFPALVAGLPIHMFRTAHNAREKLAE SLRHENLQKRESISELISLRMFLNDTLSTFDDLEKAKTHLVVLWASQANTIPATFWSLFQ MIRNPEAMKAATEEVKRTLENAGQKVSLEGNPICLSQAELNDLPVLDSIIKESLRLSSAS LNIRTAKEDFTLHLEDGSYNIRKDDIIALYPQLMHLDPEIYPDPLTFKYDRYLDENGKTK TTFYCNGLKLKYYYMPFGSGATICPGRLFAIHEIKQFLILMLSYFELELIEGQAKCPPLD QSRAGLGILPPLNDIEFKYKFKHL |
| 497 |
P35244 (RPA3) |
 Estimated TM-score=0.940 |
>Replication protein A 14 kDa subunit (Length=121) MVDMMDLPRSRINAGMLAQFIDKPVCFVGRLEKIHPTGKMFILSDGEGKNGTIELMEPLD EEISGIVEVVGRVTAKATILCTSYVQFKEDSHPFDLGLYNEAVKIIHDFPQFYPLGIVQH D |
| 498 |
P42574 (CASP3) |
 Estimated TM-score=0.940 |
>Caspase-3 (Length=277) MENTENSVDSKSIKNLEPKIIHGSESMDSGISLDNSYKMDYPEMGLCIIINNKNFHKSTG MTSRSGTDVDAANLRETFRNLKYEVRNKNDLTREEIVELMRDVSKEDHSKRSSFVCVLLS HGEEGIIFGTNGPVDLKKITNFFRGDRCRSLTGKPKLFIIQACRGTELDCGIETDSGVDD DMACHKIPVEADFLYAYSTAPGYYSWRNSKDGSWFIQSLCAMLKQYADKLEFMHILTRVN RKVATEFESFSFDATFHAKKQIPCIVSMLTKELYFYH |
| 499 |
Q9H0R4 (HDHD2) |
 Estimated TM-score=0.940 |
>Haloacid dehalogenase-like hydrolase domain-containing protein 2 (Length=259) MAACRALKAVLVDLSGTLHIEDAAVPGAQEALKRLRGASVIIRFVTNTTKESKQDLLERL RKLEFDISEDEIFTSLTAARSLLERKQVRPMLLVDDRALPDFKGIQTSDPNAVVMGLAPE HFHYQILNQAFRLLLDGAPLIAIHKARYYKRKDGLALGPGPFVTALEYATDTKATVVGKP EKTFFLEALRGTGCEPEEAVMIGDDCRDDVGGAQDVGMLGILVKTGKYRASDEEKINPPP YLTCESFPHAVDHILQHLL |
| 500 |
P02743 (APCS) |
 Estimated TM-score=0.940 |
>Serum amyloid P-component (Length=223) MNKPLLWISVLTSLLEAFAHTDLSGKVFVFPRESVTDHVNLITPLEKPLQNFTLCFRAYS DLSRAYSLFSYNTQGRDNELLVYKERVGEYSLYIGRHKVTSKVIEKFPAPVHICVSWESS SGIAEFWINGTPLVKKGLRQGYFVEAQPKIVLGQEQDSYGGKFDRSQSFVGEIGDLYMWD SVLPPENILSAYQGTPLPANILDWQALNYEIRGYVIIKPLVWV |
[an error occurred while processing this directive]
yangzhanglab![]() umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218