

You can:
| Name | Neuropeptide Y receptor type 4 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | NPY4R |
| Synonym | NPY4-R neuropeptide Y receptor type 4 pancreatic polypeptide receptor 1 PP1 Y4 receptor |
| Disease | Obesity Diabetes Schizophrenia |
| Length | 375 |
| Amino acid sequence | MNTSHLLALLLPKSPQGENRSKPLGTPYNFSEHCQDSVDVMVFIVTSYSIETVVGVLGNLCLMCVTVRQKEKANVTNLLIANLAFSDFLMCLLCQPLTAVYTIMDYWIFGETLCKMSAFIQCMSVTVSILSLVLVALERHQLIINPTGWKPSISQAYLGIVLIWVIACVLSLPFLANSILENVFHKNHSKALEFLADKVVCTESWPLAHHRTIYTTFLLLFQYCLPLGFILVCYARIYRRLQRQGRVFHKGTYSLRAGHMKQVNVVLVVMVVAFAVLWLPLHVFNSLEDWHHEAIPICHGNLIFLVCHLLAMASTCVNPFIYGFLNTNFKKEIKALVLTCQQSAPLEESEHLPLSTVHTEVSKGSLRLSGRSNPI |
| UniProt | P50391 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | P50391 |
| 3D structure model | This predicted structure model is from GPCR-EXP P50391. |
| BioLiP | N/A |
| Therapeutic Target Database | T27944 |
| ChEMBL | CHEMBL4877 |
| IUPHAR | 307 |
| DrugBank | BE0002418 |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 335 |  BDBM50442547 BDBM50442547 | C104H138N24O15 | 1964.4 | 20 / 19 | 6.9 | No |
| 7912 |  CHEMBL349328 CHEMBL349328 | C38H49IN4O2 | 720.74 | 5 / 0 | 8.2 | No |
| 8872 |  CID 73351118 CID 73351118 | C184H287N53O53S2 | 4153.76 | 61 / 58 | -13.6 | No |
| 9240 |  CID 76335489 CID 76335489 | C177H272N52O55 | 4008.43 | 62 / 60 | -15.6 | No |
| 13869 | 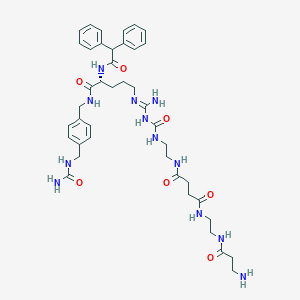 CHEMBL2440902 CHEMBL2440902 | C41H56N12O7 | 828.976 | 9 / 11 | -1.2 | No |
| 13871 | 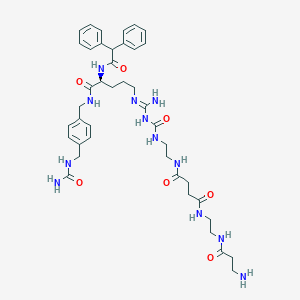 CHEMBL2440903 CHEMBL2440903 | C41H56N12O7 | 828.976 | 9 / 11 | -1.2 | No |
| 14633 | 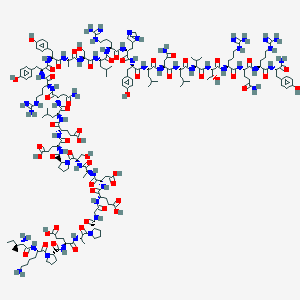 D0P9KP D0P9KP | C180H279N53O54 | 4049.53 | 61 / 61 | -14.9 | No |
| 23747 | 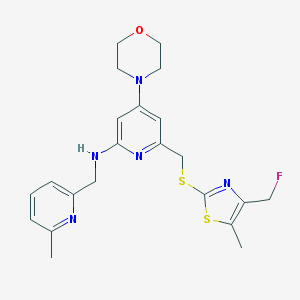 CHEMBL585711 CHEMBL585711 | C22H26FN5OS2 | 459.602 | 9 / 1 | 3.8 | Yes |
| 35085 |  CHEMBL1774211 CHEMBL1774211 | C57H65F3N8O7 | 1031.19 | 12 / 6 | N/A | No |
| 37702 |  CHEMBL3040658 CHEMBL3040658 | C84H116N18O16 | 1633.96 | 18 / 17 | 2.5 | No |
| 38213 |  CHEMBL123410 CHEMBL123410 | C22H24Cl2N2O | 403.347 | 2 / 0 | 5.4 | No |
| 38639 |  CHEMBL491762 CHEMBL491762 | C23H26O4 | 366.457 | 4 / 1 | 3.6 | Yes |
| 40668 | 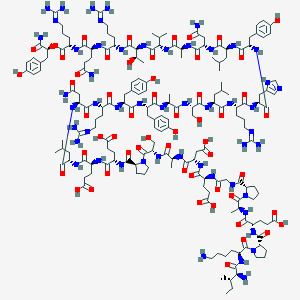 BDBM50444592 BDBM50444592 | C177H272N52O55 | 4008.43 | 62 / 56 | -17.1 | No |
| 41714 | 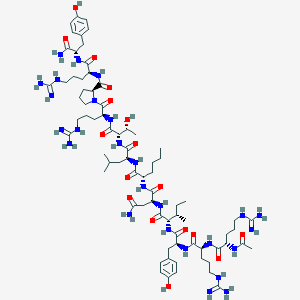 CID 72711150 CID 72711150 | C75H124N26O17 | 1661.98 | 21 / 28 | -2.5 | No |
| 468203 | 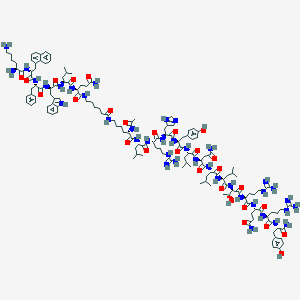 CHEMBL3578016 CHEMBL3578016 | C143H215N39O28 | 2928.54 | 34 / 41 | 2.0 | No |
| 47234 | 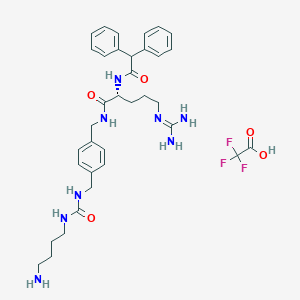 CHEMBL2441114 CHEMBL2441114 | C35H45F3N8O5 | 714.791 | 10 / 8 | N/A | No |
| 50791 | 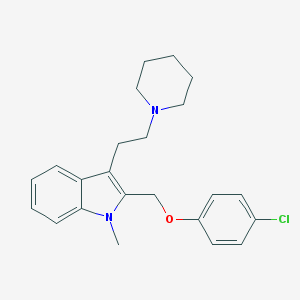 CHEMBL121799 CHEMBL121799 | C23H27ClN2O | 382.932 | 2 / 0 | 5.3 | No |
| 51682 | 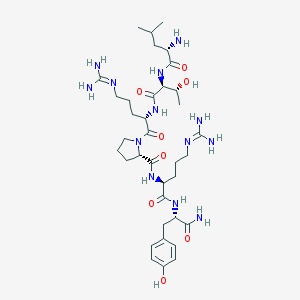 PP31-36, HUMAN PP31-36, HUMAN | C36H61N13O8 | 803.967 | 11 / 12 | -2.8 | No |
| 58515 | 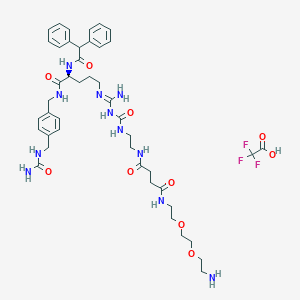 CHEMBL2440909 CHEMBL2440909 | C44H60F3N11O10 | 960.026 | 15 / 11 | N/A | No |
| 58519 | 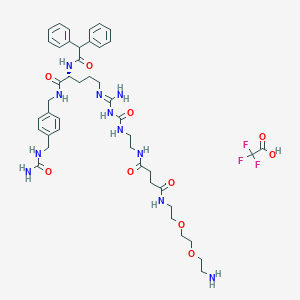 CHEMBL2440908 CHEMBL2440908 | C44H60F3N11O10 | 960.026 | 15 / 11 | N/A | No |
| 58892 | 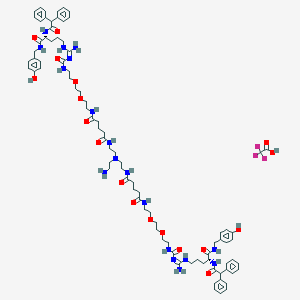 CID 73355619 CID 73355619 | C86H117F3N18O18 | 1747.98 | 23 / 18 | N/A | No |
| 59870 | 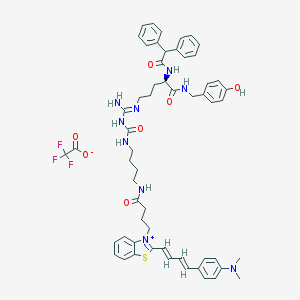 CHEMBL1774344 CHEMBL1774344 | C57H64F3N9O7S | 1076.25 | 13 / 7 | N/A | No |
| 60719 | 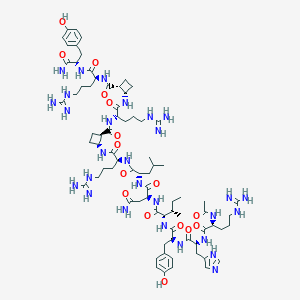 CHEMBL2440191 CHEMBL2440191 | C76H120N28O16 | 1681.98 | 21 / 29 | -2.5 | No |
| 523373 | 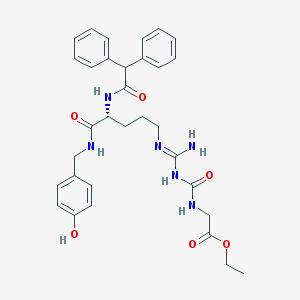 CHEMBL508974 CHEMBL508974 | C32H38N6O6 | 602.692 | 7 / 6 | 3.4 | No |
| 65000 | 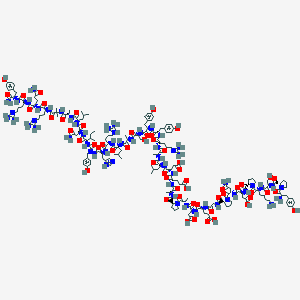 BDBM85678 BDBM85678 | C186H281N56O56+ | 4197.63 | 62 / 59 | -17.9 | No |
| 65084 | 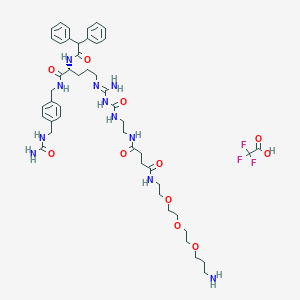 CHEMBL2440910 CHEMBL2440910 | C47H66F3N11O11 | 1018.11 | 16 / 11 | N/A | No |
| 65087 | 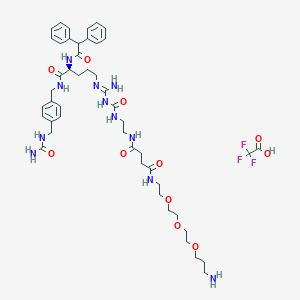 CHEMBL2440897 CHEMBL2440897 | C47H66F3N11O11 | 1018.11 | 16 / 11 | N/A | No |
| 66810 | 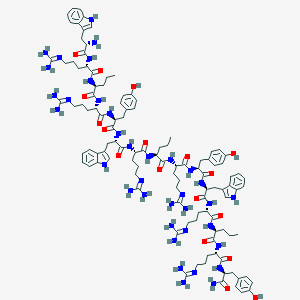 CHEMBL413871 CHEMBL413871 | C111H159N37O18 | 2299.73 | 25 / 34 | -0.6 | No |
| 68355 | 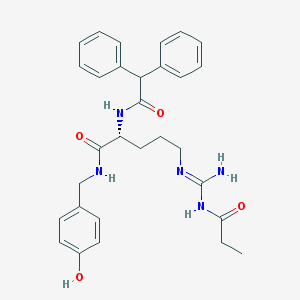 CHEMBL470495 CHEMBL470495 | C30H35N5O4 | 529.641 | 5 / 5 | 3.7 | No |
| 69889 | 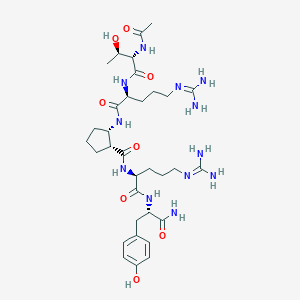 CHEMBL2440119 CHEMBL2440119 | C33H54N12O8 | 746.871 | 10 / 12 | -2.9 | No |
| 71087 | 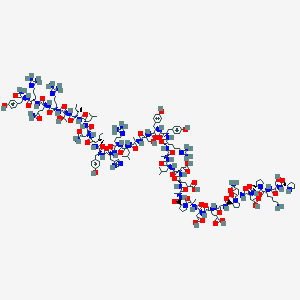 NPY2-36, porcine NPY2-36, porcine | C181H278N54O55 | 4090.54 | 62 / 58 | -17.7 | No |
| 80603 | 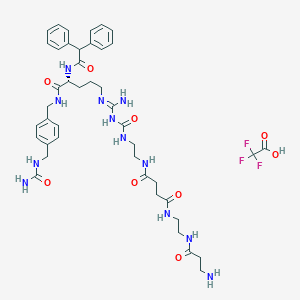 CHEMBL2440902 CHEMBL2440902 | C43H57F3N12O9 | 942.999 | 14 / 12 | N/A | No |
| 80606 | 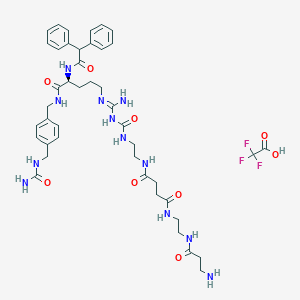 CHEMBL2440903 CHEMBL2440903 | C43H57F3N12O9 | 942.999 | 14 / 12 | N/A | No |
| 83995 | 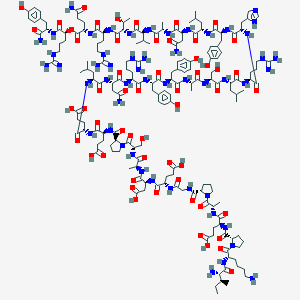 CID 76313780 CID 76313780 | C177H272N52O55 | 4008.43 | 62 / 60 | -15.6 | No |
| 84116 | 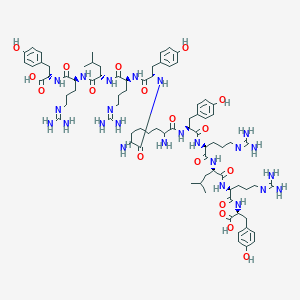 Sub[-Tyr-Arg-Leu-Arg-Tyr-NH2]2 Sub[-Tyr-Arg-Leu-Arg-Tyr-NH2]2 | C80H122N24O18 | 1708.01 | 24 / 26 | -5.3 | No |
| 84120 | 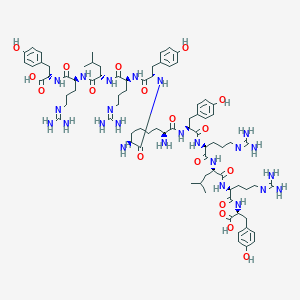 CHEMBL2371908 CHEMBL2371908 | C80H122N24O18 | 1708.01 | 24 / 26 | -5.3 | No |
| 84578 | 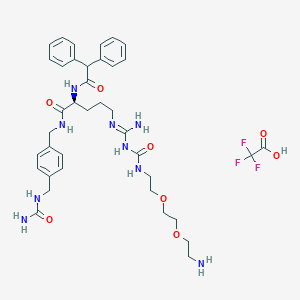 CHEMBL2440933 CHEMBL2440933 | C38H50F3N9O8 | 817.868 | 13 / 9 | N/A | No |
| 85535 | 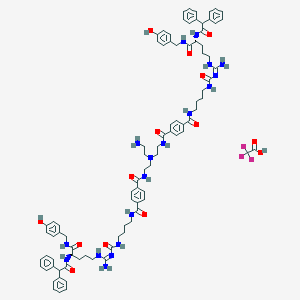 CID 73357159 CID 73357159 | C88H105F3N18O14 | 1695.92 | 19 / 18 | N/A | No |
| 86735 | 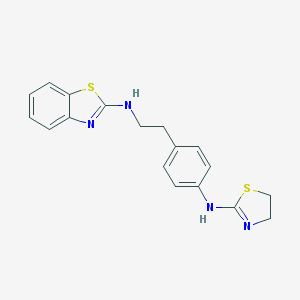 CHEMBL1081469 CHEMBL1081469 | C18H18N4S2 | 354.49 | 5 / 2 | 4.5 | Yes |
| 91261 | 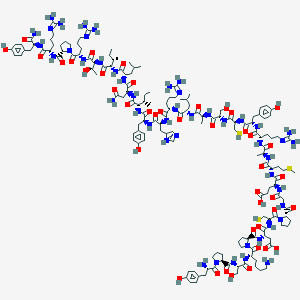 Des-AA10-17[Cys7,21, Pro34]-NPY Des-AA10-17[Cys7,21, Pro34]-NPY | C158H245N47O42S3 | 3571.17 | 52 / 49 | -12.6 | No |
| 93490 | 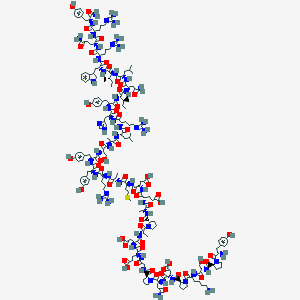 NPY D-Trp32 NPY D-Trp32 | C196H290N56O56S | 4358.87 | 65 / 59 | -15.6 | No |
| 96697 | 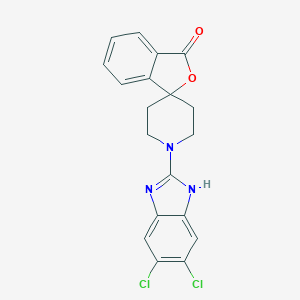 CHEMBL483251 CHEMBL483251 | C19H15Cl2N3O2 | 388.248 | 4 / 1 | 4.7 | Yes |
| 99777 | 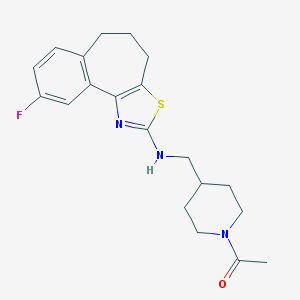 CHEMBL83255 CHEMBL83255 | C20H24FN3OS | 373.49 | 5 / 1 | 4.0 | Yes |
| 100062 | 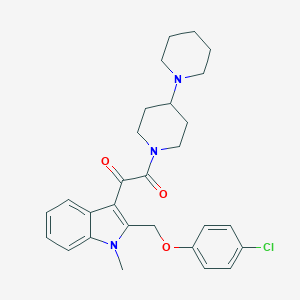 CHEMBL331202 CHEMBL331202 | C28H32ClN3O3 | 494.032 | 4 / 0 | 4.9 | Yes |
| 101497 | 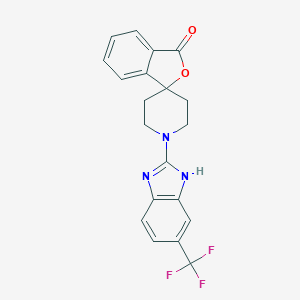 CHEMBL497359 CHEMBL497359 | C20H16F3N3O2 | 387.362 | 7 / 1 | 4.3 | Yes |
| 105856 | 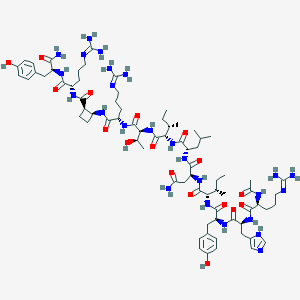 BDBM50441969 BDBM50441969 | C75H119N25O17 | 1642.94 | 21 / 24 | -2.8 | No |
| 107236 | 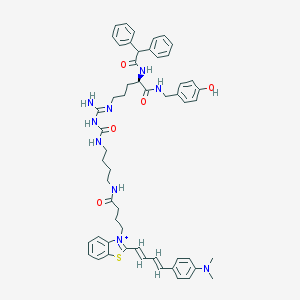 CHEMBL1774344 CHEMBL1774344 | C55H64N9O5S+ | 963.235 | 8 / 7 | 8.7 | No |
| 110613 | 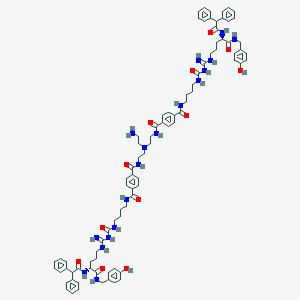 BDBM50442552 BDBM50442552 | C86H104N18O12 | 1581.89 | 16 / 19 | 7.7 | No |
| 112510 |  CID 16142386 CID 16142386 | C135H209N41O36 | 2982.41 | 42 / 48 | -6.6 | No |
| 113259 |  CHEMBL496327 CHEMBL496327 | C21H19N3O4 | 377.4 | 6 / 1 | 3.3 | Yes |
| 113378 |  BDBM50441961 BDBM50441961 | C75H124N26O17 | 1661.98 | 21 / 25 | -3.7 | No |
| 117812 |  CHEMBL2440933 CHEMBL2440933 | C36H49N9O6 | 703.845 | 8 / 8 | 0.7 | No |
| 118042 |  BDBM50441960 BDBM50441960 | C73H119N27O18 | 1662.93 | 22 / 26 | -6.6 | No |
| 124361 |  BDBM82300 BDBM82300 | C195H302N58O57S+4 | 4402.97 | 61 / 65 | -14.8 | No |
| 125588 |  BDBM50441962 BDBM50441962 | C76H126N26O17 | 1676.01 | 21 / 25 | -3.3 | No |
| 128096 |  CHEMBL2440909 CHEMBL2440909 | C42H59N11O8 | 846.003 | 10 / 10 | -0.8 | No |
| 128098 | 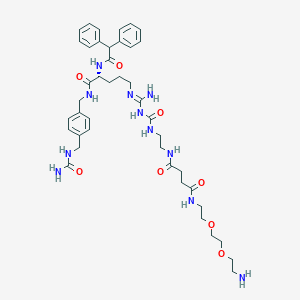 CHEMBL2440908 CHEMBL2440908 | C42H59N11O8 | 846.003 | 10 / 10 | -0.8 | No |
| 128894 | 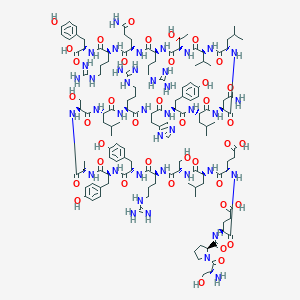 PYY13-36, porcine PYY13-36, porcine | C135H208N40O39 | 3015.39 | 45 / 49 | -8.3 | No |
| 130739 | 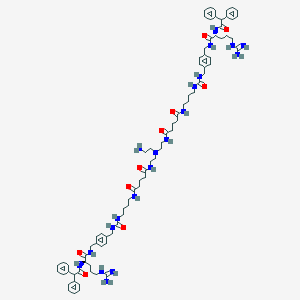 CHEMBL3040656 CHEMBL3040656 | C82H114N20O10 | 1539.94 | 14 / 19 | 2.4 | No |
| 131514 | 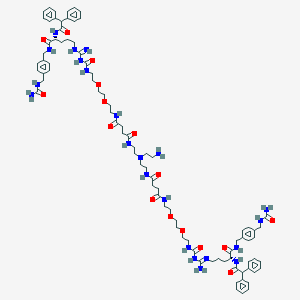 BDBM50442573 BDBM50442573 | C86H120N22O16 | 1718.04 | 20 / 20 | -0.4 | No |
| 131520 | 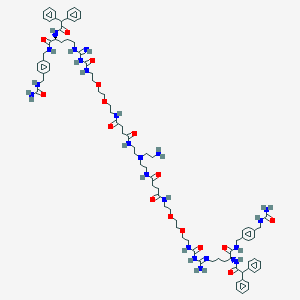 BDBM50442575 BDBM50442575 | C86H120N22O16 | 1718.04 | 20 / 20 | -0.4 | No |
| 142419 | 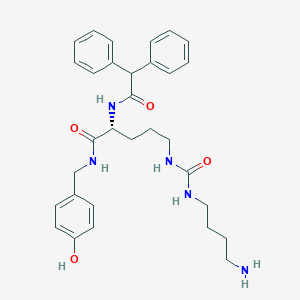 CHEMBL2440936 CHEMBL2440936 | C31H39N5O4 | 545.684 | 5 / 6 | 3.0 | No |
| 153865 | 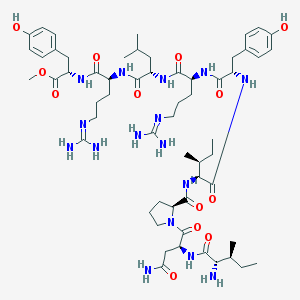 BVD 10 BVD 10 | C58H92N16O13 | 1221.47 | 16 / 15 | 0.4 | No |
| 155780 | 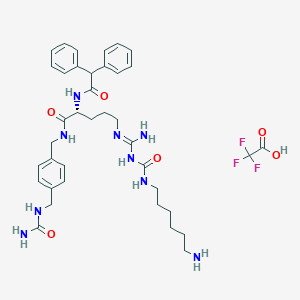 CHEMBL2440935 CHEMBL2440935 | C38H50F3N9O6 | 785.87 | 11 / 9 | N/A | No |
| 526026 | 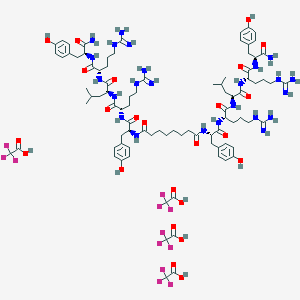 CHEMBL3785576 CHEMBL3785576 | C88H126F12N24O24 | 2132.1 | 40 / 32 | N/A | No |
| 158833 | 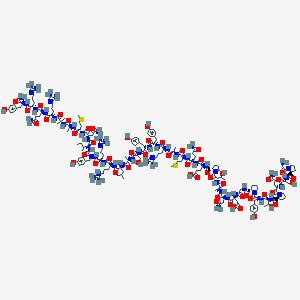 BDBM85680 BDBM85680 | C182H281N58O54S2+ | 4209.72 | 61 / 60 | -19.4 | No |
| 162271 | 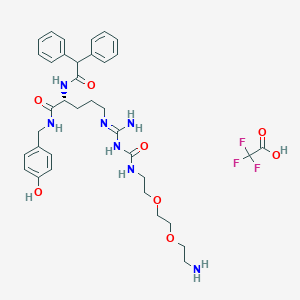 CHEMBL2440930 CHEMBL2440930 | C36H46F3N7O8 | 761.8 | 13 / 8 | N/A | No |
| 483445 | 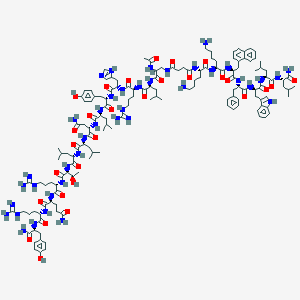 CHEMBL3578018 CHEMBL3578018 | C145H218N40O29 | 2985.59 | 35 / 42 | 1.6 | No |
| 174044 | 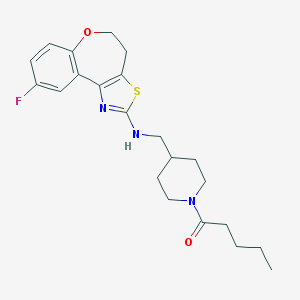 CHEMBL82374 CHEMBL82374 | C22H28FN3O2S | 417.543 | 6 / 1 | 4.7 | Yes |
| 177626 | 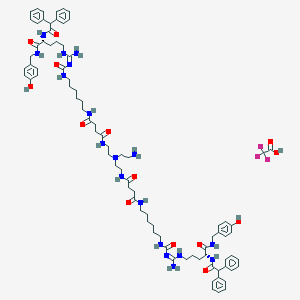 CID 73348025 CID 73348025 | C84H113F3N18O14 | 1655.94 | 19 / 18 | N/A | No |
| 178161 | 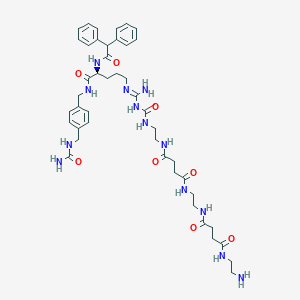 CHEMBL2440905 CHEMBL2440905 | C44H61N13O8 | 900.055 | 10 / 12 | -2.0 | No |
| 178165 | 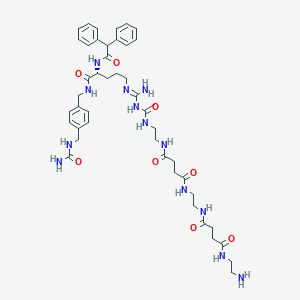 CHEMBL2440904 CHEMBL2440904 | C44H61N13O8 | 900.055 | 10 / 12 | -2.0 | No |
| 178860 | 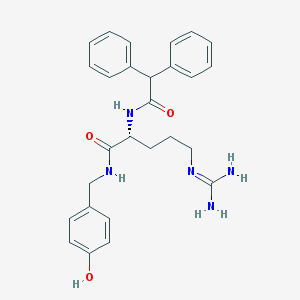 BIBP3226 BIBP3226 | C27H31N5O3 | 473.577 | 4 / 5 | 2.5 | Yes |
| 178866 | 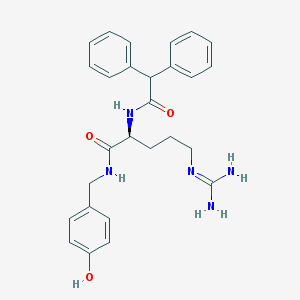 CHEMBL2440929 CHEMBL2440929 | C27H31N5O3 | 473.577 | 4 / 5 | 2.5 | Yes |
| 181566 | 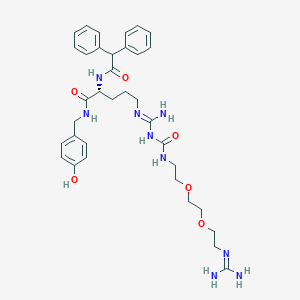 CHEMBL2440937 CHEMBL2440937 | C35H47N9O6 | 689.818 | 8 / 8 | 1.2 | No |
| 182627 | 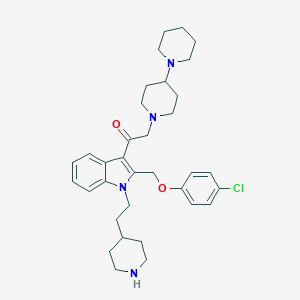 CHEMBL340369 CHEMBL340369 | C34H45ClN4O2 | 577.21 | 5 / 1 | 5.9 | No |
| 183417 |  CID 73349571 CID 73349571 | C88H121F3N22O18 | 1832.07 | 23 / 20 | N/A | No |
| 183421 |  CID 73355616 CID 73355616 | C88H121F3N22O18 | 1832.07 | 23 / 20 | N/A | No |
| 550198 |  DB05045 DB05045 | C185H288N54O55S2 | 4212.78 | 99 / 66 | 7.8 | No |
| 193056 |  BDBM85679 BDBM85679 | C184H287N53O54S2+2 | 4169.76 | 59 / 56 | -13.1 | No |
| 193926 |  CHEMBL3040691 CHEMBL3040691 | C84H116N22O12 | 1625.99 | 16 / 21 | 0.4 | No |
| 196331 |  Pim[-Trp-Arg-Nva-Arg-Tyr-NH2]2 Pim[-Trp-Arg-Nva-Arg-Tyr-NH2]2 | C81H120N28O14 | 1710.03 | 20 / 26 | -2.3 | No |
| 196335 |  CHEMBL2371910 CHEMBL2371910 | C81H120N28O14 | 1710.03 | 20 / 26 | -2.3 | No |
| 196343 |  CID 72713074 CID 72713074 | C76H123N25O17 | 1658.98 | 21 / 27 | -1.0 | No |
| 196527 | 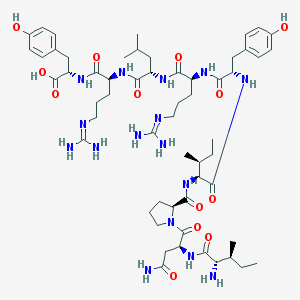 CHEMBL591971 CHEMBL591971 | C57H90N16O13 | 1207.45 | 16 / 16 | -2.2 | No |
| 196546 | 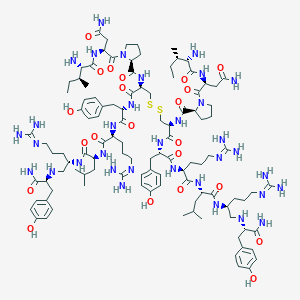 CID 44283143 CID 44283143 | C108H172N34O22S2 | 2362.9 | 32 / 32 | -3.6 | No |
| 198328 | 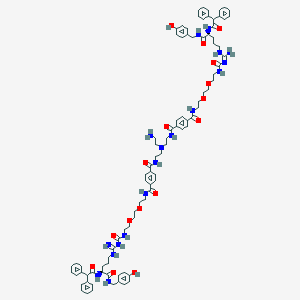 CID 73443032 CID 73443032 | C90H112N18O16 | 1702.0 | 19 / 18 | 5.5 | No |
| 198333 | 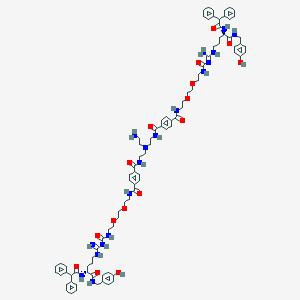 CID 73443030 CID 73443030 | C90H112N18O16 | 1702.0 | 19 / 18 | 5.5 | No |
| 199252 | 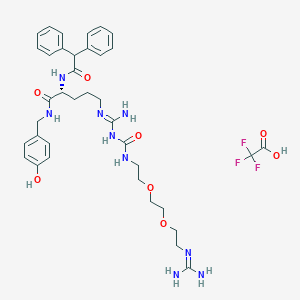 CHEMBL2440937 CHEMBL2440937 | C37H48F3N9O8 | 803.841 | 13 / 9 | N/A | No |
| 200027 | 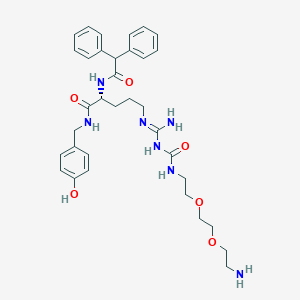 CHEMBL2440930 CHEMBL2440930 | C34H45N7O6 | 647.777 | 8 / 7 | 1.8 | No |
| 202950 | 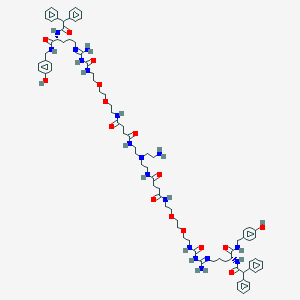 BDBM50442557 BDBM50442557 | C82H112N18O16 | 1605.91 | 20 / 17 | 1.4 | No |
| 204561 | 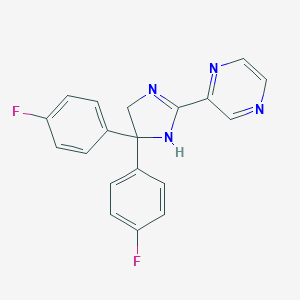 CHEMBL490937 CHEMBL490937 | C19H14F2N4 | 336.346 | 5 / 1 | 2.3 | Yes |
| 207674 | 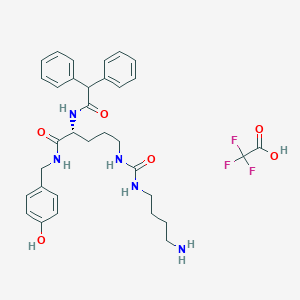 CHEMBL2440936 CHEMBL2440936 | C33H40F3N5O6 | 659.707 | 10 / 7 | N/A | No |
| 207962 | 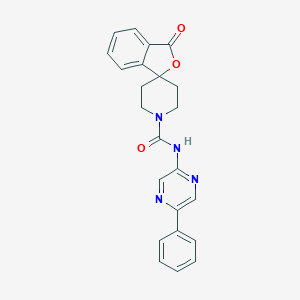 CHEMBL491930 CHEMBL491930 | C23H20N4O3 | 400.438 | 5 / 1 | 2.4 | Yes |
| 209126 | 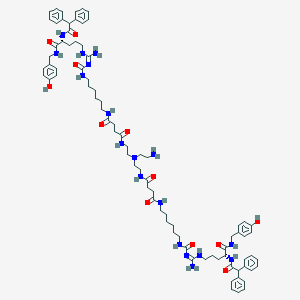 CHEMBL3040675 CHEMBL3040675 | C82H112N18O12 | 1541.91 | 14 / 17 | 5.2 | No |
| 212980 | 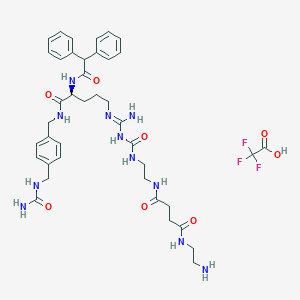 CHEMBL2440901 CHEMBL2440901 | C40H52F3N11O8 | 871.92 | 13 / 11 | N/A | No |
| 212982 | 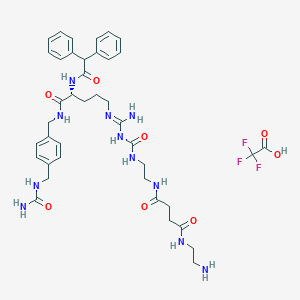 CHEMBL2440900 CHEMBL2440900 | C40H52F3N11O8 | 871.92 | 13 / 11 | N/A | No |
| 213655 | 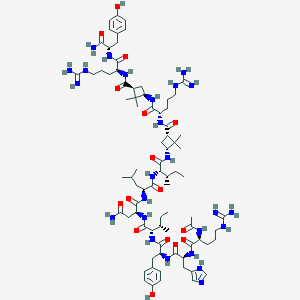 CID 73354074 CID 73354074 | C80H127N25O16 | 1695.06 | 20 / 26 | 0.2 | No |
| 214651 | 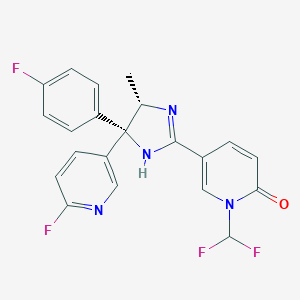 CHEMBL1784130 CHEMBL1784130 | C21H16F4N4O | 416.38 | 7 / 1 | 2.9 | Yes |
| 490040 | 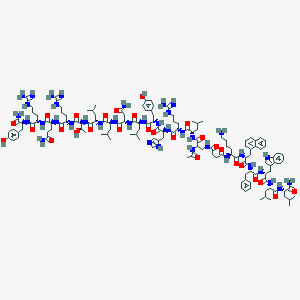 CHEMBL3578017 CHEMBL3578017 | C139H206N38O28 | 2857.42 | 33 / 40 | 2.1 | No |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417