

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MNTSHLLALLLPKSPQGENRSKPLGTPYNFSEHCQDSVDVMVFIVTSYSIETVVGVLGNLCLMCVTVRQKEKANVTNLLIANLAFSDFLMCLLCQPLTAVYTIMDYWIFGETLCKMSAFIQCMSVTVSILSLVLVALERHQLIINPTGWKPSISQAYLGIVLIWVIACVLSLPFLANSILENVFHKNHSKALEFLADKVVCTESWPLAHHRTIYTTFLLLFQYCLPLGFILVCYARIYRRLQRQGRVFHKGTYSLRAGHMKQVNVVLVVMVVAFAVLWLPLHVFNSLEDWHHEAIPICHGNLIFLVCHLLAMASTCVNPFIYGFLNTNFKKEIKALVLTCQQSAPLEESEHLPLSTVHTEVSKGSLRLSGRSNPI | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCCCCCSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 987654212588999888878888999876545568716898899999999999988999998999708988838899999999999999999899999999838677966889889999999999999999999998586732768631116568886469999999999999999750100246765433357987444066894689999999999999999999999999999999802578887514589888355588999999999999948999999999964454416699999999999999999999999997599999999999510046898777888788887768889988888888999 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MNTSHLLALLLPKSPQGENRSKPLGTPYNFSEHCQDSVDVMVFIVTSYSIETVVGVLGNLCLMCVTVRQKEKANVTNLLIANLAFSDFLMCLLCQPLTAVYTIMDYWIFGETLCKMSAFIQCMSVTVSILSLVLVALERHQLIINPTGWKPSISQAYLGIVLIWVIACVLSLPFLANSILENVFHKNHSKALEFLADKVVCTESWPLAHHRTIYTTFLLLFQYCLPLGFILVCYARIYRRLQRQGRVFHKGTYSLRAGHMKQVNVVLVVMVVAFAVLWLPLHVFNSLEDWHHEAIPICHGNLIFLVCHLLAMASTCVNPFIYGFLNTNFKKEIKALVLTCQQSAPLEESEHLPLSTVHTEVSKGSLRLSGRSNPI | |
| 534231222023323433232333333232243233221000000211120021023112100000001340210001001000300100010001010000003201102000100000000000000000000000002000000233213310000000011100000100000020232334433331433332000101012730220000010221113101000200000001013334336554444334432000000000000000011120000001001332243200100102000000100010000000004301400240011023434455454343333334334334434446437 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCCCCCSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MNTSHLLALLLPKSPQGENRSKPLGTPYNFSEHCQDSVDVMVFIVTSYSIETVVGVLGNLCLMCVTVRQKEKANVTNLLIANLAFSDFLMCLLCQPLTAVYTIMDYWIFGETLCKMSAFIQCMSVTVSILSLVLVALERHQLIINPTGWKPSISQAYLGIVLIWVIACVLSLPFLANSILENVFHKNHSKALEFLADKVVCTESWPLAHHRTIYTTFLLLFQYCLPLGFILVCYARIYRRLQRQGRVFHKGTYSLRAGHMKQVNVVLVVMVVAFAVLWLPLHVFNSLEDWHHEAIPICHGNLIFLVCHLLAMASTCVNPFIYGFLNTNFKKEIKALVLTCQQSAPLEESEHLPLSTVHTEVSKGSLRLSGRSNPI | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.26 | 0.23 | 0.79 | 3.03 | Download | ----------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLM--TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD------------GAVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS---KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------------------- | |||||||||||||||||||
| 2 | 4n6hA | 0.27 | 0.23 | 0.77 | 3.97 | Download | ------------------------------------SLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALA-TSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTRPR------------DGAVVCMLQFPSPSWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS---KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKP----------------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.26 | 0.23 | 0.79 | 3.65 | Download | ----------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATST-LPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD------------GAVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSG---SKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------------------- | |||||||||||||||||||
| 4 | 4djh | 0.24 | 0.21 | 0.76 | 1.54 | Download | ------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVT-TTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDTPLKAKIINICIWLLSSSVGISAIVLGGTKVRE----------DVDVIECSLQFPDDDWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNIPYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------------------------------------ | |||||||||||||||||||
| 5 | 4rnb | 0.28 | 0.28 | 0.77 | 1.15 | Download | ------------------------------------PKEYEWVLIAGYIIVFVVALIGNVLVCVAVWKNHHMRTVTNYFIVNLSLADVLVTITCLPATLVVDITETWFFGQSLCKVIPYLQTVSVSVSVLTLSCIALDRWYAICHPST----AKRARNSIVIIWIVSCIIMIPQAIVMECSTVFKT---------TLFTVCDERWGGEIYPKMYHICFFLVTYMAPLCLMVLAYLQIFRKLWCRQGIDCSSSFSKQIRARRKTARMLMVVLLVFAICYLPISILNVLKRVFGMFHDRETVYAWFTFSHWLVYANSAANPIIYNFLSGKFREEFKAAFSC------------------------------------ | |||||||||||||||||||
| 6 | 4n6hA | 0.26 | 0.23 | 0.79 | 3.26 | Download | ------------------------------GARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALA-TSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD------------GAVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSG---SKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------------------- | |||||||||||||||||||
| 7 | 3uon | 0.21 | 0.20 | 0.75 | 1.70 | Download | ----------------------------------------VVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVTTKMAGMMIAAAWVLSFILWAPAILFWQFIVGVRTEDGE----------CYIQFFSN---AAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIFGAWDAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPCI----PNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM------------------------------------- | |||||||||||||||||||
| 8 | 4ea3A | 0.26 | 0.22 | 0.74 | 4.75 | Download | ------------------------------------PLGLKVTIVGLYLAVCVGGLLGNCLVMYVILRHTKMKTATNIYIFNLALADTLVLL-TLPFQGTDILLGFWPFGNALCKTVIAIDYYNMFTSTFTLTAMSVDRYVAICHPT-----SSKAQAVNVAIWALASVVGVPVAIMGSAQVEDEEI----------ECLVEIPTPQDYWGPVFAICIFLFSFIVPVLVISVCYSLMIRRLRGVRLLSGS---REKDRNLRRITRLVLVVVAVFVGCWTPVQVFVLAQGLGVQP-SSETAVAILRFCTALGYVNSCLNPILYAFLDENFKACFR----------------------------------------- | |||||||||||||||||||
| 9 | 4n6hA | 0.26 | 0.23 | 0.79 | 3.08 | Download | ----------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLM--TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTRPR------------DGAVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS---KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------------------- | |||||||||||||||||||
| 10 | 5c1mA | 0.24 | 0.22 | 0.78 | 3.71 | Download | -------------------------GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVRYTKMKTATNIYIFNLALADALAT-STLPFQSVNYLMGTWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKAFRTPRNAKIVNVCNWILSSAIGLPVMFMATTKYR------------QGSIDCTLTFPTWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMLSGS---KEKDRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKALI-TIPETTFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF------------------------------------------ | |||||||||||||||||||
| ||||||||||||||||||||||||||
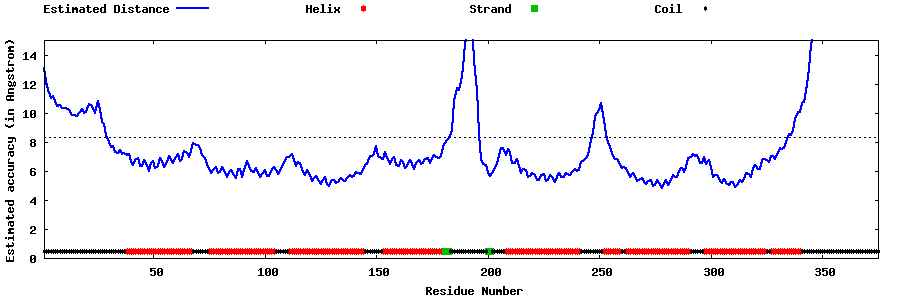
| Generated 3D models | Estimated local accuracy of models | ||
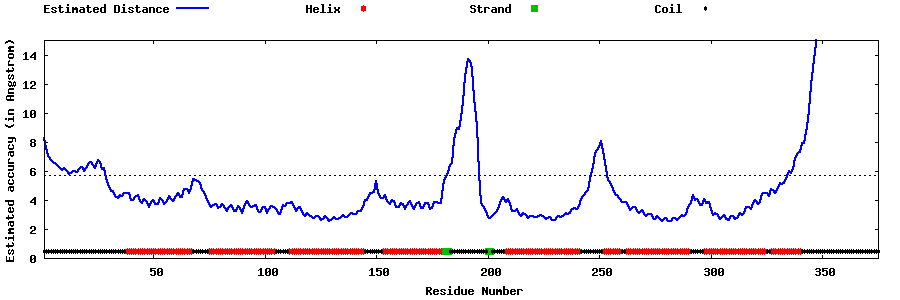 | |||
|
|
|||
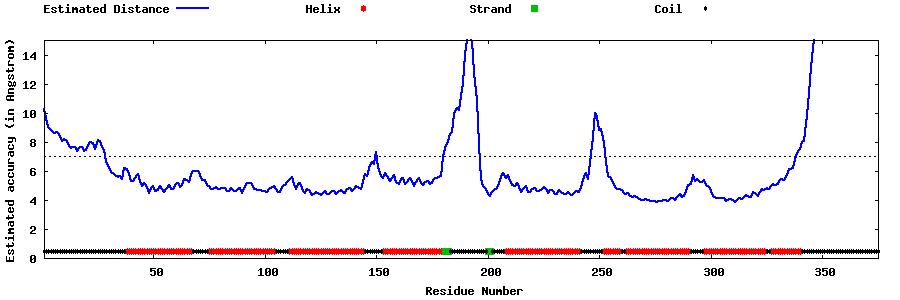 | |||
|
| |||
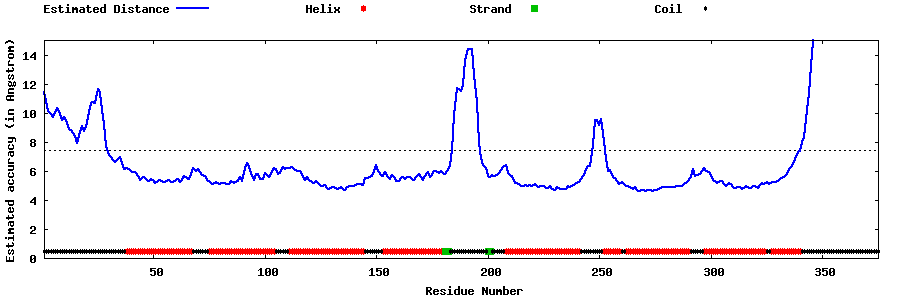 | |||
|
| |||
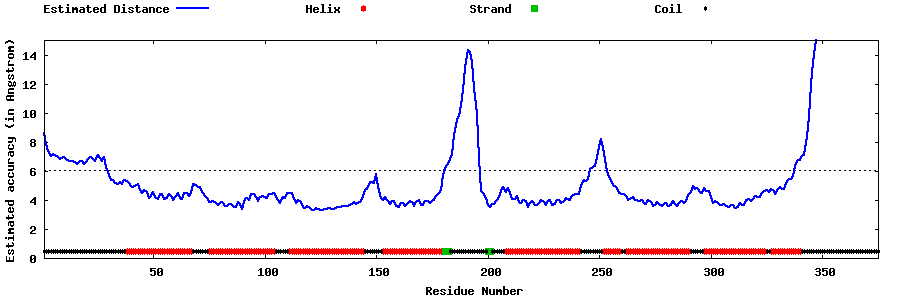 | |||
|
| |||
 | |||
|
| |||