

You can:
| Name | 5-hydroxytryptamine receptor 2A |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Htr2a |
| Synonym | serotonin 5HT-2 receptor 5Ht-2 'D' receptor 5-hydroxytryptamine (serotonin) receptor 2A, G protein-coupled 5-HT2A receptor [ Show all ] |
| Disease | N/A for non-human GPCRs |
| Length | 471 |
| Amino acid sequence | MEILCEDNISLSSIPNSLMQLGDGPRLYHNDFNSRDANTSEASNWTIDAENRTNLSCEGYLPPTCLSILHLQEKNWSALLTTVVIILTIAGNILVIMAVSLEKKLQNATNYFLMSLAIADMLLGFLVMPVSMLTILYGYRWPLPSKLCAIWIYLDVLFSTASIMHLCAISLDRYVAIQNPIHHSRFNSRTKAFLKIIAVWTISVGISMPIPVFGLQDDSKVFKEGSCLLADDNFVLIGSFVAFFIPLTIMVITYFLTIKSLQKEATLCVSDLSTRAKLASFSFLPQSSLSSEKLFQRSIHREPGSYAGRRTMQSISNEQKACKVLGIVFFLFVVMWCPFFITNIMAVICKESCNENVIGALLNVFVWIGYLSSAVNPLVYTLFNKTYRSAFSRYIQCQYKENRKPLQLILVNTIPALAYKSSQLQVGQKKNSQEDAEQTVDDCSMVTLGKQQSEENCTDNIETVNEKVSCV |
| UniProt | P14842 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL322 |
| IUPHAR | 6 |
| DrugBank | N/A |
| Name | Ketanserin |
|---|---|
| Molecular formula | C22H22FN3O3 |
| IUPAC name | 3-[2-[4-(4-fluorobenzoyl)piperidin-1-yl]ethyl]-1H-quinazoline-2,4-dione |
| Molecular weight | 395.434 |
| Hydrogen bond acceptor | 5 |
| Hydrogen bond donor | 1 |
| XlogP | 2.6 |
| Synonyms | KETANSERIN, Ketanserin tartrate hydrate, KETANSERIN TARTRATE 3-[2-[4-(4-fluorobenzoyl)piperidin-1-yl]ethyl]-1H-quinazoline-2,4-dione KS-00000XLH 74050-98-9 NCGC00021146-04 [ Show all ] |
| Inchi Key | FPCCSQOGAWCVBH-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C22H22FN3O3/c23-17-7-5-15(6-8-17)20(27)16-9-11-25(12-10-16)13-14-26-21(28)18-3-1-2-4-19(18)24-22(26)29/h1-8,16H,9-14H2,(H,24,29) |
| PubChem CID | 3822 |
| ChEMBL | CHEMBL51 |
| IUPHAR | 197, 88 |
| BindingDB | 21395 |
| DrugBank | DB12465 |
Structure | 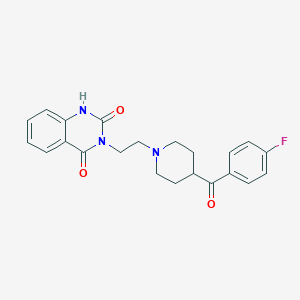 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| IC50 | 3.4 nM | PMID8960552 | BindingDB,ChEMBL |
| Kd | 0.2818 nM | Bioorg. Med. Chem. Lett., (1995) 5:7:667 | ChEMBL |
| Kd | 1.349 nM | PMID11754579 | ChEMBL |
| Kd | 1.35 nM | PMID11754579 | BindingDB |
| Kd | 1.58489 nM | PMID11040033 | IUPHAR |
| Ki | 0.47 nM | PMID24050112 | ChEMBL |
| Ki | 0.49 nM | PMID17900912 | BindingDB,ChEMBL |
| Ki | 0.75 nM | PMID1347569 | BindingDB |
| Ki | 0.85 nM | PMID27061981, PMID26820556 | BindingDB |
| Ki | 0.85 nM | PMID19954866, PMID22133459, PMID27061981, PMID21440338, PMID26820556 | BindingDB,ChEMBL |
| Ki | 0.89 nM | PMID9225287 | BindingDB |
| Ki | 1.0 nM | PMID7984267 | BindingDB |
| Ki | 1.0 - 2.51189 nM | PMID11040033, PMID9655845 | IUPHAR |
| Ki | 1.04 nM | PMID6645787 | BindingDB |
| Ki | 1.07 nM | PMID6645787 | BindingDB |
| Ki | 1.2 nM | PMID2795135 | BindingDB |
| Ki | 1.25 nM | PMID7582481 | BindingDB |
| Ki | 1.29 nM | PMID6645787 | BindingDB |
| Ki | 1.5 nM | PMID11354383 | BindingDB,ChEMBL |
| Ki | 1.6 nM | PMID9686407 | BindingDB |
| Ki | 1.99 nM | PMID7984267 | BindingDB |
| Ki | 3.2 nM | PMID2877462 | BindingDB |
| Ki | 3.5 nM | PMID17649988 | BindingDB,ChEMBL |
| Ki | 3.98 nM | PMID7984267 | BindingDB |
| Ki | 5.01 nM | PMID7984267 | BindingDB |
| Ki | 5.129 nM | PMID11754579 | ChEMBL |
| Ki | 14.5 nM | PMID1501121 | BindingDB |
| Ki | 192.0 nM | PMID2531826 | BindingDB |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417