| Synonyms | SVR
Bayer 205
C51H40N6O23S6
CTK7G6721
DTXSID1046344
Harnstoff, symmetischer der 3-aminobenzoyl-3-amino-4-methylbenzoyl-1-naphthylamin-4,6,8-trisulfonsaeure
1,3,5-Naphthalenetrisulfonic acid, 8,8'-[carbonylbis[imino-3,1-phenylenecarbonylimino(4-methyl-3,1-phenylene)carbonylimino]]bis-(9CI)
KBioSS_002402
1,5-Naphthylenetrisulfonic acid, 8,8'-[ureylenebis[m-phenylenecarbonylimino(4-methyl-m-phenylene)carbonylimino]]di-, hexasodium salt
Naganil
5,5'',5''''-[1,3,6-naphthalenetriyltris(sulfonylimino)]tris[1,3-benzenesulfonate analogue
NCGC00163318-02
8,8'-[Carbonylbis[imino-3,1-phenylenecarbonylimino(4-methyl-3,1-phenylene)carbonyl-imino]]bis-1,3,5-napthalenetrisulfonic acid
SpecPlus_000662
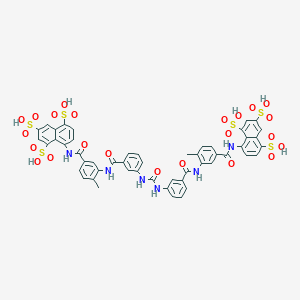
8-[(4-methyl-3-{[3-({[3-({2-methyl-5-[(4,6,8-trisulfonaphthalen-1-yl)carbamoyl]phenyl}carbamoyl)phenyl]carbamoyl}amino)benzene]amido}benzene)amido]naphthalene-1,3,5-trisulfonic acid
8-{4-methyl-3-[3-({[3-({2-methyl-5-[(4,6,8-trisulfonaphthalen-1-yl)carbamoyl]phenyl}carbamoyl)phenyl]carbamoyl}amino)benzamido]benzamido}naphthalene-1,3,5-trisulfonic acid
Tox21_112045_1
BRD-K70327167-348-01-9
CCG-36173
DB04786
Farma 939
KBio2_002397
1,3,5-Naphthalenetrisulfonic acid, 8,8'-[ureylenebis[m-phenylenecarbonylimino(4-methyl-m-phenylene)carbonylimino]]di-, hexasodium salt
LS-95867
104283-EP2301627A1
Naganol
8,8''-(Carbonylbis(imino-3,1-phenylenecarbonylimino(4-methyl-3,1-phenylene)carbonylimino))bisnaphthalene-1,3,5-trisulphonic acid
NSC-34936
8,8'-[carbonylbis[imino-3,1-phenylenecarbonylimino(4-methyl-3,1-phenylene)carbonylimino]]bis-1,3,5-naphthalenetrisulphonic acid
Spectrum_001877
AN-43641
WLN: L66J BSWQ DSWQ GSWQ JMVR D1 CMVR CMVMR CVMR B1 EVM-JL66J BSWQ DSWQ GSWQ &-NA-6
BSPBio_002248
CHEMBL265502
DSSTox_GSID_46344
Germanin (TN)
1,3,5-naphthalenetrisulfonic acid, 8,8'-[carbonylbis[imino-3,1-phenylenecarbonylimino(4-methyl-3,1-phenylene)carbonylimino]]bis-
KBio3_001468
1,5-Naphthalenetrisulfonic acid, 8,8'-[carbonylbis[imino-3,1-phenylenecarbonylimino(4-methyl-3,1-phenylene)carbonylimino]]bis-, hexasodium salt
Moranil
NCGC00025177-08
8,8'-(Carbonylbis(imino-3,1-phenylenecarbonylimino(4-methyl-3,1-phenylene)carbonylimino))bisnaphthalene-1,3,5-trisulphonic acid
SGCTO-001
8,8'-{carbonylbis[imino-3,1-phenylenecarbonylimino(4-methyl-3,1-phenylene)carbonylimino]}dinaphthalene-1,3,5-trisulfonic acid
Suramine
sym-bis(m-aminobenzoyl-m-amino-p-methylbenzoyl-1-napthylamino-4,6,8-trisulfonate) carbamide
BDBM50336799
Carbanilide, 3,3'-bis((5-((4,6,8-trisulfo-1-naphthyl)carbamoyl)-o-tolyl)carbamoyl)-
D0H2UB
EINECS 205-658-4
HY-B0879
1,3,5-Naphthalenetrisulfonic acid, 8,8'-[ureylenebis[m-phenylenecarbonylimino(4-methyl-m-phenylene)carbonylimino]]di- (8CI)
L000585
1-(3-Benzamido-4-methylbenzamido)naphthalene-4,8-trisulfonic acid sym-3''-urea sodium salt
Naganin
NCGC00163318-03
8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFONIC
Spectrum3_000724
8-[[4-methyl-3-[[3-[[3-[[2-methyl-5-[(4,6,8-trisulfo-1-naphthyl)carbamoyl]phenyl]carbamoyl]phenyl]carbamoylamino]benzoyl]amino]benzoyl]amino]naphthalene-1,3,5-trisulfonic acid
AC1L1K6K
UNII-6032D45BEM
BRD-K70327167-348-09-2
CHEBI:45906
DivK1c_006758
Fourneau
KBio2_004965
1,3,5-Naphthalenetrisulfonicacid,8,8'-[carbonylbis[imino-3,1-phenylenecarbonylimino(4-methyl-3,1-phenylene)carbonylimino]]bis-
m,m'-Ureylen-bis-(8-(m-(benzamido)-p-toluamido)naphthalin-1,3,5-trisulfonsaeure)
129-46-4 (hexa-hydrochloride salt)
Naganol 6 Na
8,8''-[Carbonylbis[imino-3,1-phenylenecarbonylimino(4-methyl-3,1-phenylene)carbonyl-imino]]bis-1,3,5-naphthalenetrisulfonic acid(suramin)
NSC34936
8,8'-[carbonylbis[imino-3,1-phenylenecarbonylimino(4-methyl-3,1-phenylene)carbonylimino]]bisnaphthalene-1,3,5-trisulphonic acid
Suramine sodium
Antrypol
C07974
CS-4444
DSSTox_RID_81548
GTPL1728
1,3,5-Naphthalenetrisulfonic acid, 8,8'-[carbonylbis[imino-3,1-phenylenecarbonylimino(4-methyl-3,1-phenylene)carbonylimino]]bis- (9CI)
KBioGR_001774
1,5-Naphthalenetrisulfonic acid, 8,8'-[ureylenebis[m-phenylenecarbonylimino(4-methyl-m-phenylene)carbonylimino]]di-, hexasodium salt
Moranyl
3-14-00-02267 (Beilstein Handbook Reference)
NCGC00163318-01
8,8'-[Carbonylbis[imino-3,1-phenylenecarbonylimino(4-methyl-3,1-phenylene)carbonyl-imino]]bis-1,3,5-naphthalenetrisulfonic acid
Sodium suramin
8,8-[Carbonylbis[imino-3,1-phenylenecarbonylimino(4-methyl-3,1-phenylene)carbonylimino]]bisnaphthalene-1,3,5-trisulphonic acid
Tox21_112045
Belganyl
CAS-145-63-1
Farma
KBio1_001702
1,3,5-Naphthalenetrisulfonic acid, 8,8'-[ureylenebis[m-phenylenecarbonylimino(4-methyl-m-phenylene)carbonylimino]]di-(8CI)
Lopac0_001182
104283-EP2298761A1
Naganine
6032D45BEM
NSC 34936
8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFONIC ACID
Spectrum4_001247
8-[[4-methyl-3-[[3-[[3-[[2-methyl-5-[(4,6,8-trisulfonaphthalen-1-yl)carbamoyl]phenyl]carbamoyl]phenyl]carbamoylamino]benzoyl]amino]benzoyl]amino]naphthalene-1,3,5-trisulfonic acid
AKOS030526554
US8835659, Suramin
BRN 3230873
DSSTox_CID_26344
Germanin
1,3,5-Naphthalenetrisulfonic acid, 8,8'-(carbonylbis(imino-3,1-phenylenecarbonylimino(4-methyl-3,1-phenylene)carbonylimino))bis-
KBio2_007533
1,3,5-Naphthylenetrisulfonic acid, 8,8'-(ureylenebis(m-phenylenecarbonylimino(4-methyl-m-phenylene)carbonylimino))di-
MMV637953
145-63-1
Naphuride
8,8''-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFONIC ACID
SCHEMBL3161
8,8'-[Ureylenebis[m-phenylenecarbonylimino(4-methyl-m-phenylene)carbonylimino]]di-1,3,5-naphthalenetrisulfonic acid
Suramin, Hexasodium Salt [ Show all ] |
|---|


![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417