

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MAANVSGAKSCPANFLAAADDKLSGFQGDFLWPILVVEFLVAVASNGLALYRFSIRKQRPWHPAVVFSVQLAVSDLLCALTLPPLAAYLYPPKHWRYGEAACRLERFLFTCNLLGSVIFITCISLNRYLGIVHPFFARSHLRPKHAWAVSAAGWVLAALLAMPTLSFSHLKRPQQGAGNCSVARPEACIKCLGTADHGLAAYRAYSLVLAGLGCGLPLLLTLAAYGALGRAVLRSPGMTVAEKLRVAALVASGVALYASSYVPYHIMRVLNVDARRRWSTRCPSFADIAQATAALELGPYVGYQVMRGLMPLAFCVHPLLYMAAVPSLGCCCRHCPGYRDSWNPEDAKSTGQALPLNATAAPKPSEPQSRELSQ | |
| CCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCSSSSCCCCCHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 97856899899876445555323656688389999999999999899976666652588998999999999999999999709999999668999997088999999999999999999999999978999813010014162468777799999999999999997221665899547840476212210244320169999999999999999999999999999999984377877431117999999999999999579999999999987524555214554423235677889999999999999999785999992388898999985243435787767887762489877789999998877779 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MAANVSGAKSCPANFLAAADDKLSGFQGDFLWPILVVEFLVAVASNGLALYRFSIRKQRPWHPAVVFSVQLAVSDLLCALTLPPLAAYLYPPKHWRYGEAACRLERFLFTCNLLGSVIFITCISLNRYLGIVHPFFARSHLRPKHAWAVSAAGWVLAALLAMPTLSFSHLKRPQQGAGNCSVARPEACIKCLGTADHGLAAYRAYSLVLAGLGCGLPLLLTLAAYGALGRAVLRSPGMTVAEKLRVAALVASGVALYASSYVPYHIMRVLNVDARRRWSTRCPSFADIAQATAALELGPYVGYQVMRGLMPLAFCVHPLLYMAAVPSLGCCCRHCPGYRDSWNPEDAKSTGQALPLNATAAPKPSEPQSRELSQ | |
| 75434143442343323135441440211000210120023023200200000001233433000000033100201000000010000024431100100010010111210100000000001000000000020242232200000000010000000100000020344753311011212442243242343322021011112123013211200020000000001315446443220000000000000120133112110010013132244143343133333323310200110010001200021000000004501410240033244344543544444344534434454466354358 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCSSSSCCCCCHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MAANVSGAKSCPANFLAAADDKLSGFQGDFLWPILVVEFLVAVASNGLALYRFSIRKQRPWHPAVVFSVQLAVSDLLCALTLPPLAAYLYPPKHWRYGEAACRLERFLFTCNLLGSVIFITCISLNRYLGIVHPFFARSHLRPKHAWAVSAAGWVLAALLAMPTLSFSHLKRPQQGAGNCSVARPEACIKCLGTADHGLAAYRAYSLVLAGLGCGLPLLLTLAAYGALGRAVLRSPGMTVAEKLRVAALVASGVALYASSYVPYHIMRVLNVDARRRWSTRCPSFADIAQATAALELGPYVGYQVMRGLMPLAFCVHPLLYMAAVPSLGCCCRHCPGYRDSWNPEDAKSTGQALPLNATAAPKPSEPQSRELSQ | |||||||||||||||||||||||||
| 1 | 4mbsA | 0.21 | 0.20 | 0.79 | 3.15 | Download | ----------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINY-KRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEG-LHYTCSSHFPYS---------QYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEEEK-KRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNN-----------CSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------------- | |||||||||||||||||||
| 2 | 4mbsA | 0.22 | 0.20 | 0.79 | 4.24 | Download | -----------------CQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILI-NYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAA--AQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQK-EGLHYTCSSHFPY---------SQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMEVEEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNC-----------SSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------------- | |||||||||||||||||||
| 3 | 4mbsA | 0.21 | 0.20 | 0.79 | 3.58 | Download | ----------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINY-KRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGL-HYTCSSHFPY---------SQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEEE-KKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNN-----------CSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------------- | |||||||||||||||||||
| 4 | 4ib4 | 0.17 | 0.21 | 0.76 | 1.51 | Download | ---------------------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVS-LEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIFEAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIKGIETN-------------PNNITCVLTKE----RFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCN-----------------QTTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR--------------------------------- | |||||||||||||||||||
| 5 | 4djh | 0.18 | 0.21 | 0.75 | 1.16 | Download | -----------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVI-IRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLM-NSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRDVDVIECSLQFPDD---------DYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLEKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGS----------------------AALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------------------------------------ | |||||||||||||||||||
| 6 | 5vblB | 0.23 | 0.24 | 0.80 | 3.30 | Download | -------------------EYTDWKSSGALIPAIYMLVFLLGTTGNGLVLWTVFRSSREKRRSADIFIASLAVADLTFVVTLPLWATYTYRDYDWPFGTFFCKLSSYLIFVNMYASAFCLTGLSFDRYLAIVRPVANARLRLRVSGAVATAVLWVLAALLAMPVMVLRTTGDLENTKVQCYMDYSMVATV-----SSEWAWEVGLGVSSTTVGFVVPFTIMLTCYFFIAQTIAE-------ERRRLLSIIVVLVVTFALCKMPYHLVKTLYMLGSLLHWP------------CDFDLFLMNIFPYCTCISYVNSCLNPFLYAFFDPRFRQACTSMLLMGQSR-------------------------------- | |||||||||||||||||||
| 7 | 3uon | 0.17 | 0.26 | 0.76 | 1.69 | Download | ---------------------------VVFIVLVAGSLSLVTIIGNILVMVSIKV-NRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIVGTVEDGECYIQFFSNAA--V----TFGTAIAAFYLPVIIMTVLYWHISRASKSINIEGGGTWDAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAP----------------C----IPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM------------------------------------- | |||||||||||||||||||
| 8 | 4mbsA | 0.22 | 0.19 | 0.78 | 4.21 | Download | ----------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILI-NYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGL-HYTCSSHFP---------YSQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRE-----KKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNN-----------CSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------------- | |||||||||||||||||||
| 9 | 5vblB | 0.23 | 0.24 | 0.80 | 3.38 | Download | -----------------CEYTDW-KSSGALIPAIYMLVFLLGTTGNGLVLWTVFRSSREKRRSADIFIASLAVADLTFVVTLPLWATYTYRDYDWPFGTFFCKLSSYLIFVNMYASAFCLTGLSFDRYLAIVRPVANARLRLRVSGAVATAVLWVLAALLAMPVMVLRTTGDLENNKVQCYMDYSM-----VATVSSEWAWEVGLGVSSTTVGFVVPFTIMLTCYFFIAQTIAE-------ERRRLLSIIVVLVVTFALCKMPYHLVKTLYMLGSLLHWP------------CDFDLFLMNIFPYCTCISYVNSCLNPFLYAFFDPRFRQACTSMLLMGQSR-------------------------------- | |||||||||||||||||||
| 10 | 5c1mA | 0.18 | 0.18 | 0.78 | 3.71 | Download | ------------GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVR-YTKMKTATNIYIFNLALADALATSTLPFQSVNYLMG-TWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRTPRNAKIVNVCNWILSSAIGLPVMFMATTKY-RQGSIDCTLTFSHP----------TWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMLSGSKERRITRMVLVVVAVFIVCWTPIHIYVIIKALITIPET-----------------TFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF------------------------------------------ | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
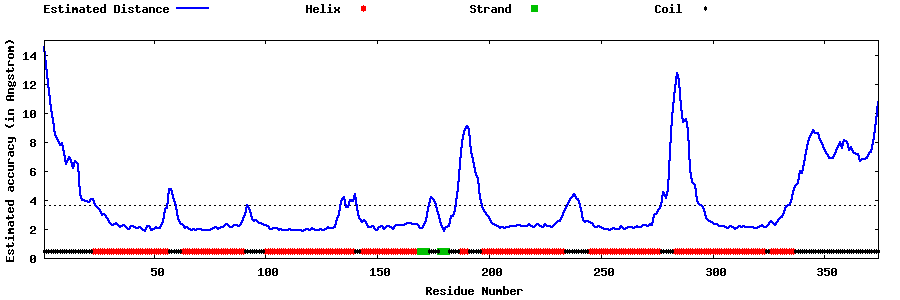 | |||
|
|
|||
 | |||
|
| |||
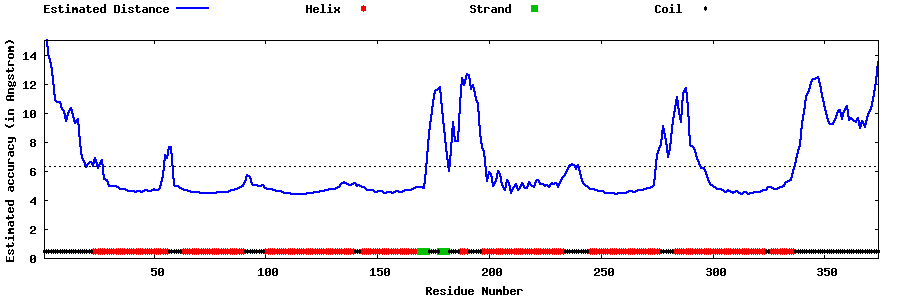 | |||
|
| |||
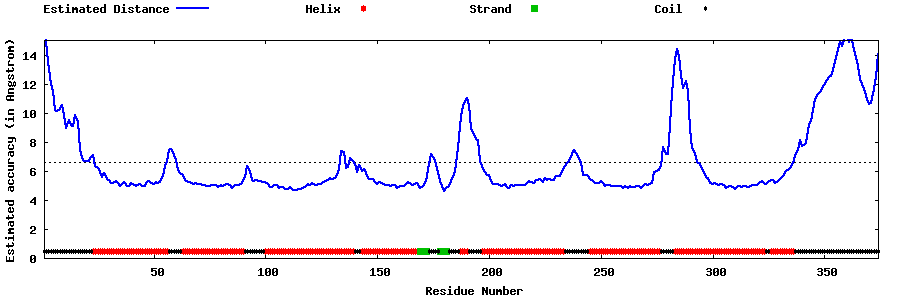 | |||
|
| |||
 | |||
|
| |||