

You can:
| Name | 8-Cyclopentyl-1,3-dipropylxanthine |
|---|---|
| Molecular formula | C16H24N4O2 |
| IUPAC name | 8-cyclopentyl-1,3-dipropyl-7H-purine-2,6-dione |
| Molecular weight | 304.394 |
| Hydrogen bond acceptor | 3 |
| Hydrogen bond donor | 1 |
| XlogP | 4.0 |
| Synonyms | PD 116948 BDBM21173 SC-91856 CCG-39732 Spectrum3_001073 [ Show all ] |
| Inchi Key | FFBDFADSZUINTG-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C16H24N4O2/c1-3-9-19-14-12(15(21)20(10-4-2)16(19)22)17-13(18-14)11-7-5-6-8-11/h11H,3-10H2,1-2H3,(H,17,18) |
| PubChem CID | 1329 |
| ChEMBL | CHEMBL183 |
| IUPHAR | 386 |
| BindingDB | 21173 |
| DrugBank | N/A |
Structure | 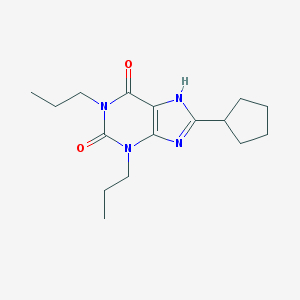 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| ED50 | <10.0 uM | PMID8676354 | ChEMBL |
| IC50 | 217.0 nM | PMID10476879 | BindingDB,ChEMBL |
| IC50 | 261.0 nM | PMID10476879 | BindingDB,ChEMBL |
| Kd | 30.0 nM | PMID27282729 | BindingDB |
| Kd | 76.29 nM | DOI: http://dx.doi.org/10.6019/CHEMBL3885741 | ChEMBL |
| Kd | 106.44 nM | DOI: http://dx.doi.org/10.6019/CHEMBL3885741 | ChEMBL |
| Ki | 56.0 nM | PMID16250647 | BindingDB,ChEMBL |
| Ki | 63.0957 - 251.189 nM | PMID9459566, PMID9933143, PMID9920286, PMID9179373, PMID16902942 | IUPHAR |
| Ki | 70.0 nM | PMID10476879 | BindingDB,ChEMBL |
| Ki | 129.0 nM | PMID18258439, J Med Chem. 2005 Mar 24;48(6):2045-53., PMID23200243, PMID15771447, PMID9459566 | PDSP,BindingDB,ChEMBL |
| Ki | 130.0 nM | PMID20408530, PMID18637670, PMID19036477, PMID19282184 | BindingDB,ChEMBL |
| Ki | 144.0 nM | PMID15163184 | BindingDB,ChEMBL |
| Ki | 156.0 nM | PMID11809867 | PDSP,BindingDB |
| Ki | 157.0 nM | PMID26824742 | BindingDB,ChEMBL |
| Ki | 200.0 nM | PMID15163184 | BindingDB,ChEMBL |
| Ki | 226.0 nM | PMID9258366 | BindingDB,ChEMBL |
| Ki | 260.0 nM | PMID19301821, PMID20937560, PMID17665891, PMID25462223, PMID16789747, PMID18468446, PMID16335918 | BindingDB,ChEMBL |
| Ki | 260.3 nM | PMID25462223 | ChEMBL |
| Ki | 291.0 nM | PMID9258366 | BindingDB,ChEMBL |
| Ki | 330.0 nM | PMID2825043 | BindingDB |
| Ki | 337.0 nM | PMID17927167, PMID15214785, PMID16789747, PMID24077183, PMID18269230, PMID22257095, PMID11462973, PMID16366607, PMID10956220, PMID27282729, PMID16335918 | BindingDB,ChEMBL |
| Ki | 337.0 nM | PMID10956220 | BindingDB |
| Ki | 340.0 nM | PMID8071944 | BindingDB,ChEMBL |
| Ki | 630.0 nM | PMID23200243 | ChEMBL |
| Ki | 788.0 nM | PMID9258366 | BindingDB,ChEMBL |
| koff | 0.4596 min^-1 | DOI: http://dx.doi.org/10.6019/CHEMBL3885741 | ChEMBL |
| kon | 0.00306 nM^-1 min^-1 | DOI: http://dx.doi.org/10.6019/CHEMBL3885741 | ChEMBL |
| kon | 451467.0 Ms-1 | DOI: http://dx.doi.org/10.6019/CHEMBL3885741 | ChEMBL |
| kon | 482900.0 Ms-1 | DOI: http://dx.doi.org/10.6019/CHEMBL3885741 | ChEMBL |
| k_off | 0.03484 s-1 | DOI: http://dx.doi.org/10.6019/CHEMBL3885741 | ChEMBL |
| k_off | 0.04447 s-1 | DOI: http://dx.doi.org/10.6019/CHEMBL3885741 | ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417