

You can:
| Name | Adenosine receptor A1 |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Adora1 |
| Synonym | A1 receptor A1-AR A1R adenosine receptor A1 RDC7 |
| Disease | N/A for non-human GPCRs |
| Length | 326 |
| Amino acid sequence | MPPYISAFQAAYIGIEVLIALVSVPGNVLVIWAVKVNQALRDATFCFIVSLAVADVAVGALVIPLAILINIGPQTYFHTCLMVACPVLILTQSSILALLAIAVDRYLRVKIPLRYKTVVTQRRAAVAIAGCWILSLVVGLTPMFGWNNLSVVEQDWRANGSVGEPVIKCEFEKVISMEYMVYFNFFVWVLPPLLLMVLIYLEVFYLIRKQLNKKVSASSGDPQKYYGKELKIAKSLALILFLFALSWLPLHILNCITLFCPTCQKPSILIYIAIFLTHGNSAMNPIVYAFRIHKFRVTFLKIWNDHFRCQPKPPIDEDLPEEKAED |
| UniProt | P25099 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL318 |
| IUPHAR | 18 |
| DrugBank | N/A |
| Name | 8-Cyclopentyl-1,3-dipropylxanthine |
|---|---|
| Molecular formula | C16H24N4O2 |
| IUPAC name | 8-cyclopentyl-1,3-dipropyl-7H-purine-2,6-dione |
| Molecular weight | 304.394 |
| Hydrogen bond acceptor | 3 |
| Hydrogen bond donor | 1 |
| XlogP | 4.0 |
| Synonyms | [DPCPX] I14-10012 KBioGR_001696 MLS000069347 102146-07-6 [ Show all ] |
| Inchi Key | FFBDFADSZUINTG-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C16H24N4O2/c1-3-9-19-14-12(15(21)20(10-4-2)16(19)22)17-13(18-14)11-7-5-6-8-11/h11H,3-10H2,1-2H3,(H,17,18) |
| PubChem CID | 1329 |
| ChEMBL | CHEMBL183 |
| IUPHAR | 386 |
| BindingDB | 21173 |
| DrugBank | N/A |
Structure | 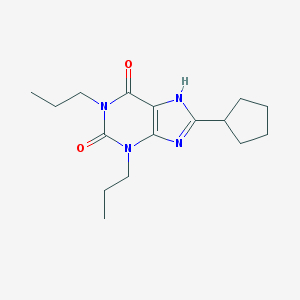 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| Activity | 86.0 % | PMID12398547 | ChEMBL |
| IC50 | 4.7 nM | , PMID10450966, Bioorg. Med. Chem. Lett., (1996) 6:17:2059 | BindingDB,ChEMBL |
| IC50 | 6.0 nM | PMID12398547 | BindingDB,ChEMBL |
| Kb | 0.23 nM | PMID1548682 | ChEMBL |
| Kb | 0.9 nM | PMID3346878 | ChEMBL |
| Kd | 5.754 nM | PMID9871584 | ChEMBL |
| Ki | 0.18 nM | PMID2067592 | BindingDB |
| Ki | 0.18 - 1.0 nM | PMID16902942, PMID12014951, PMID2067592 | IUPHAR |
| Ki | 0.211 nM | PMID26392370 | ChEMBL |
| Ki | 0.211 nM | PMID26392370 | BindingDB |
| Ki | 0.229 nM | PMID26392370 | BindingDB |
| Ki | 0.229 nM | PMID26392370 | ChEMBL |
| Ki | 0.23 nM | PMID2016719, PMID1613758 | BindingDB,ChEMBL |
| Ki | 0.23 nM | PMID1613758 | BindingDB |
| Ki | 0.2818 nM | PMID10072675 | ChEMBL |
| Ki | 0.3 nM | PMID26392370, PMID2825043 | BindingDB,ChEMBL |
| Ki | 0.3 nM | PMID26392370 | BindingDB |
| Ki | 0.45 nM | PMID2825043 | BindingDB |
| Ki | 0.46 nM | PMID1501234 | BindingDB |
| Ki | 0.46 nM | PMID1738138, PMID1613758, PMID8355252, PMID7932565, PMID1501234, PMID1548682 | BindingDB,ChEMBL |
| Ki | 0.477 nM | PMID26392370 | BindingDB |
| Ki | 0.477 nM | PMID26392370 | ChEMBL |
| Ki | 0.49 nM | PMID1501234 | BindingDB,ChEMBL |
| Ki | 0.5 nM | PMID17602796, PMID20116907 | BindingDB,ChEMBL |
| Ki | 0.553 nM | PMID26392370 | BindingDB |
| Ki | 0.553 nM | PMID26392370 | ChEMBL |
| Ki | 0.56 nM | PMID26392370 | ChEMBL |
| Ki | 0.56 nM | PMID26392370 | BindingDB |
| Ki | 0.6 nM | PMID2724296 | BindingDB |
| Ki | 0.6 nM | PMID2724296 | ChEMBL |
| Ki | 0.6166 nM | PMID1542091 | ChEMBL |
| Ki | 0.62 nM | PMID1542091 | BindingDB |
| Ki | 0.77 nM | PMID11960496 | BindingDB,ChEMBL |
| Ki | 0.9 nM | PMID15771453, PMID1613758, PMID1501234, PMID2724296, PMID11906291, PMID2754711, PMID20188574 | BindingDB,ChEMBL |
| Ki | 0.9 nM | PMID2754711 | BindingDB |
| Ki | 0.9 nM | PMID1501234, PMID1613758, PMID20188574 | BindingDB |
| Ki | 1.0 nM | PMID12014951 | BindingDB,ChEMBL |
| Ki | 1.2 nM | PMID11960496 | BindingDB,ChEMBL |
| Ki | 1.46 nM | PMID11960496 | BindingDB,ChEMBL |
| Ki | 1.5 nM | PMID8676354 | BindingDB,ChEMBL |
| Ki | 3.77 nM | PMID9191953 | BindingDB,ChEMBL |
| Ki | 340.0 nM | PMID2825043 | BindingDB |
| Ratio Ki | 0.86 - | PMID26392370 | ChEMBL |
| Ratio Ki | 0.92 - | PMID26392370 | ChEMBL |
| Ratio Ki | 1.0 - | PMID26392370 | ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417