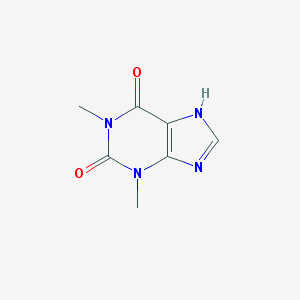
| Synonyms | NCGC00018117-06
Uniphyllin continus
Duraphyl
NCGC00018117-14
Xanthine,3-dimethyl-
Elixophyllin
NCGC00022112-03
Euphylong
NCGC00022112-12
HMS2092M05
NSC-757346
I14-16783
Optiphyllin
KBioGR_000785
Physpan
Lanophyllin
Prestwick3_000873
MFCD00079619
Theostat
Purine-2,6(1H,3H)-dione, 1,3-dimethyl-
Mudrane
Uni-Dur
Diffumal
Aerobin
Theobid Jr.
SBI-0050003.P004
AI3-50216
Theoclear-200
1,3-dimethyl-1,3,7-trihydropurine-2,6-dione
Slo-phyllin
Asmalix
Theodur G
1,3-dimethyl-3,9-dihydro-1H-purine-2,6-dione
Somophyllin-CRT
BIDD:GT0151
Theolix
1H-purine-2,6-dione
Spectrum2_000842
Bronchoparat
Theophyline
SR-01000075195-3
C07130
Theophylline aminoacetate
6461-EP2272844A1
Talotren
CHEMBL190
Theophylline [USP]
6461-EP2301936A1
Teofilina [Polish]
component of Primatene
Theophylline, Pharmaceutical Secondary Standard; Certified Reference Material
8-(2-Furyl)theophylline
Theal tablets
component of Theo-organdin (Salt/Mix)
AB00052106_24
Theo-dur
CS-4158
Quibron-T
Doraphyllin
NCGC00018117-09
UPCMLD-DP123:001
Egifilin
NCGC00018117-17
Z271004650
Elixophyllin(e)
NCGC00022112-07
Glycine Theophyllinate
NCI60_001736
HMS3369N14
Nuelin
KBio1_000203
PDSP1_001234
KS-0000477R
Prestwick1_000820
Lopac0_000014
PubChem9695
MLS002153487
Theovent
NCGC00018117-01
UNII-0I55128JYK
D00371
Respbid
Aerolate SR
Theoclair-SR
SCHEMBL4915
AN-23602
Theocontin
1,3-dimethyl-2,3,6,7-tetrahydro-1H-purine-2,6-dione
SMR000058537
BBL023514
Theograd
1,3-Dimethylxanthine
SomophyllinT
BPBio1_001041
Theonite
1H-Purine-2,6-dione, 3,9-dihydro-1,3-dimethyl-
Spectrum5_001232
Bronkodyl SR
Theophylline (1,3-dimethylxanthine)
56645-32-0
STK397040
CCG-20301
Theophylline Anhydrous,(S)
6461-EP2281815A1
Telbans
component of Bronkotabs (Salt/Mix)
Theophylline, anhydrous
6461-EP2305640A2
Teonova
component of Quibron (Salt/Mix)
Theoplus
A3133D49-AAB6-49A1-B60B-B5198F327D3F
theo von ct
component of Theolair plus (Salt/Mix)
Quibron T
AB1009249
Theo-Organidin
NCGC00018117-04
Uniphyl (TN)
DSSTox_RID_76090
NCGC00018117-12
X 115
Elixicon
NCGC00018117-20
Euphyllin
NCGC00022112-10
HMS1921E03
NSC 2066
HY-B0809
Nuelin SA
KBio2_006654
Pharmakon1600-01500568
LaBID
Prestwick2_000873
MCULE-8236489917
Purine,6(1H,3H)-dione, 1,3-dimethyl-
MolPort-001-002-058
Tox21_202375
Theobid Duracap
RP24142
Afonilum
Theoclear L.A.-130
0I55128JYK
Slo Phyllin
ARONIS25346
Theodur
1,3-Dimethyl-3,7-dihydro-1H-purine-2,6-dione
Somophyllin CRT
BDBM82053
Theolair (TN)
111079-49-3
SPBio_002866
BRD-K97799481-002-03-6
Theophyl-225
3,7-dihydro-1,3-dimethyl-1H- purine-2,6-dione
SR-01000075195
BSPBio_000945
Theophylline (JP17)
6461-EP2272835A1
Synophylate-L.A. Cenules
CHEBI:28177
theophylline solu-tion
6461-EP2298305A1
Teocen 200
component of Hecadrol (Salt/Mix)
Theophylline, certified reference material, United States Pharmacopeia (USP) Reference Standard
6461-EP2314590A1
Theacitin
component of Slo-phyllin GG (Salt/Mix)
AB00052106-22
Theo-24 (TN)
ConstantT
Quibron T/SR
Accurbron
NCGC00018117-07
Uniphylline
Duraphyllin
NCGC00018117-15
Xanthium
Elixophyllin (TN)
NCGC00022112-04
F2173-0145
NCGC00254040-01
HMS2233E16
NSC2066
IDI1_000203
Parkophyllin
KBioSS_001518
Prestwick0_000820
LASMA
Pro-vent
MLS000069390
Theostat 80
Q-201819
N1442
Unicontin CR
Dimethylxanthine
Aerolate
Theochron
SC-05534
AKOS000120961
Theoclear-80
1,3-dimethyl-1H-purine-2,6(3H,7H)-dione
SloPhyllin
Asmax
Theodur G (TN)
1,3-Dimethyl-6-hydroxy-1,3-dihydro-2H-purin-2-one
Somophyllin-DF
Bilordyl
Theolixir
1H-Purine-2,6-dione, 3,7-dihydro-1,3-dimethyl-
Spectrum3_000672
Bronchoretard
Theophyllin
46157-00-0
SR-01000075195-5
C7H8N4O2
Theophylline anhydrous
6461-EP2280006A1
Tefamin
Choledyl SA
Theophylline, 99+%, anhydrous
6461-EP2301937A1
Teofyllamin
component of Primatene (Salt/Mix)
Theophylline-SR
Theo 24
component of Theo-Organidin
AB00052106_25
Theo-Dur-Sprinkle
ct, theo von
Quibron-t (TN)
Unilong
DSSTox_CID_1336
NCGC00018117-10
von ct, theo
EINECS 200-385-7
NCGC00018117-18
ZFXYFBGIUFBOJW-UHFFFAOYSA-N
Elixophylline
NCGC00022112-08
GS 2591A
NE11194
HMS500K05
Nuelin S.A
KBio2_001518
PDSP2_001002
KSC491S8L
Prestwick1_000873
LS-241
Pulmidur
MLS004491910
Tox21_110827
NCGC00018117-02
D0F8RA
Respicur
Aescin-IIA
Theoclear
(8alpha, 9S)-10,11-Dihydro-6'-methoxycinchonan-9-amine trihydrochloride, min. 90%
SCHEMBL8312163
Aquaphyllin
Theodel
1,3-Dimethyl-2,6-dioxo-1,2,3,6-tetrahydropurine
SMR003435989
BCBcMAP01_000071
Theokin
1024160-72-2
SPBio_000823
BRD-K97799481-001-02-0
Theopek
2,6-Dihydroxy-1,3-dimethylpurine
Spectrum_001038
Bronkotabs
Theophylline (anhydrous)
58-55-9
Sustaire
CCRIS 4729
Theophylline melting point standard, Pharmaceutical Secondary Standard; Certified Reference Material
6461-EP2286811A1
Telbans Dry Syrup
component of Dicurin Procaine
Theophylline, anhydrous, >=99%, powder
6461-EP2308562A2
Teosona
component of Quibron Plus (Salt/Mix)
Theospan
AB00052106
Theo-11
Constant T
Quibron T SR
AC-20328
Theo24
NCGC00018117-05
Uniphyllin
DTXSID5021336
NCGC00018117-13
Xanthine, 1,3-dimethyl-
Elixomin
NCGC00018117-23
Euphylline
NCGC00022112-11
HMS2089A06
NSC-2066
Hylate
Opera_ID_76
KBio3_001583
Physpa
Labophylline
Prestwick3_000820
Medaphyllin
Purine-2,3H)-dione, 1,3-dimethyl-
MolPort-001-737-342
Tox21_300028
DB00277
Acet-theocin
THEOBID JR
SAM002554935
Afonilum Retard
Theoclear LA
1,3 Dimethylxanthine
Slo-bid
Asbron
Theodur Dry Syrup
1,3-Dimethyl-3,7-dihydro-1H-purine-2,6-dione #
Somophyllin T
BIDD:ER0557
Theolair-SR
1H-Purine-2, 3,7-dihydro-1,3-dimethyl-
SPECTRUM1500568
Bronchodid Duracap
Theophyl-SR
3,7-Dihydro-1,3-dimethyl-1H-purine-2,6-dione
SR-01000075195-1
BSPBio_002363
Theophylline 1.0 mg/ml in Methanol
6461-EP2272841A1
T-Phyl
Theophylline solution, 1.0 mg/mL in methanol, ampule of 1 mL, certified reference material
6461-EP2301933A1
Teofilina
component of Mudrane GG elixir (Salt/Mix)
Theophylline, European Pharmacopoeia (EP) Reference Standard
75448-53-2
Theal tabl.
component of Tedral (Salt/Mix)
AB00052106-23
Theo-DS
CPD000058537
Quibron TSR
DivK1c_000203
NCGC00018117-08
UPCMLD-DP123
EC 200-385-7
NCGC00018117-16
Xantivent
Elixophyllin SR
NCGC00022112-05
FT-0631259
NCGC00259924-01
HMS3259O06
NSC757346
InChI=1/C7H8N4O2/c1-10-5-4(8-3-9-5)6(12)11(2)7(10)13/h3H,1-2H3,(H,8,9
PDSP1_001018
KS-000010T0
Prestwick0_000873
Liquophylline
Pseudotheophylline
MLS002152943
Theotard
NC00542
Unifyl
Quibron-T/SR
Aerolate III
Theocin
SC-95891
AKOS005434016
Theoconfin Continuous
1,3-dimethyl-1H-purine-2,6(3H,9H)-dione
SMP1_000291
Austyn
Theofol
1,3-dimethyl-7H-purine-2,6-dione
Somophyllin-t
BPBio1_000791
Theona P
1H-Purine-2,6-dione, 3,7-dihydro-1,3-dimethyl-, mo
Spectrum4_000353
Bronkodyl
4eoh
ST024762
CAS-58-55-9
Theophylline Anhydrous, USP
6461-EP2281813A1
Telb-DS
Chronophyllin
Theophylline, >=99.0% (HPLC)
6461-EP2305219A1
Teolair
component of Primatene tablets (Salt/Mix)
Theophylline-[8-3H
83377-EP2371823A1
Theo Dur
component of Theo-Organidin (Salt/Mix)
Quadrinal
AB00052106_26
Theo-Nite
CTK3J1985
Uniphyl
DSSTox_GSID_21336
NCGC00018117-11
WLN: T56 BM DN FNVNVJ F1 H1
Elixex
NCGC00018117-19
ZINC18043251
Etheophyl
NCGC00022112-09
GTPL413
NINDS_000203
HSDB 3399
Nuelin S.A.
KBio2_004086
PDSP2_001218
L000595
Prestwick2_000820
Maphylline
Pulmo-Timelets
MLS006011970
Tox21_110827_1
NCGC00018117-03
DB-053224
Respid
Afonilm
Theoclear 80
SDCCGMLS-0066620.P001
Armophylline
Theodrip
1,3-Dimethyl-2,6-dioxo-1,2,3,6-tetrahydropurine; 1,3-Dimethylxanthine; 3,7-Dihydro-1,3-dimethyl-1H-purine-2,6-dione
Solosin
BDBM10847
Theolair
SPBio_002640
BRD-K97799481-001-12-9
Theophyl
2a3a
Spophyllin retard
BSPBio_000719
Theophylline (JP15)
6461-EP1441224A2
Synophylate
Cetraphylline
Theophylline melting point standard, United States Pharmacopeia (USP) Reference Standard
6461-EP2295434A2
Telbans DrySyrup
component of Dicurin Procaine (Salt/Mix)
Theophylline, British Pharmacopoeia (BP) Reference Standard
6461-EP2311827A1
Tesona
component of Quibron-T/SR (Salt/Mix)
AB00052106-20
Theo-24
Constant-T
Quibron T-SR
AC1L1D1F
Theobid [ Show all ] |
|---|


![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417