| Synonyms | Trazepam
AC1L1EZE
Diastat Acudial
Valitran
Anlin
Diazepam (intranasal, epilepsy)
Vazen
Apo-diazepam
Diazepam for system suitability, European Pharmacopoeia (EP) Reference Standard
Wy 3467
Baogin
Diazepam [USAN:USP:INN:BAN:JAN]
Betapam
S.A.R.L.
C-17931
Serenack
11100-37-1
Caudel
Simasedan
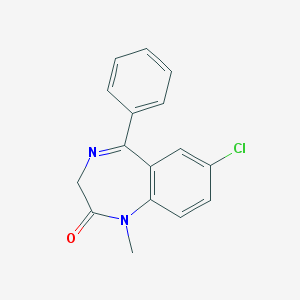
7 -chloro-1,3-dihydro-1-methyl-5-phenyl-2H-1,4-benzodiazepin-2-one
Stesolid
7-chloro-1-methyl-5-phenyl-1,3-dihydro-benzo[e][1,4]diazepin-2-one
D07JVL
Tox21_202458
7-Chloro-1-methyl-5-phenyl-3H-1,4-benzodiazepin-2(1H)-one
Dialag
Oprea1_126223
Gradual
Paxel
HY-17027
Pomin
KBio3_002780
Quiatril
La-Iii
Ro 5-2807
Mandro
Diazepamum
Methyldiazepinone
Dipaz
NC00247
DSSTox_CID_406
Nervium
DZP
Novazam
Faustal
Diapax
Unisedil
Aliseum
Diazapam
Valium R
Ansiolisina
Diazepam 1.0 mg/ml in Methanol
Vival
Arzepam
Diazepam Rectubes
Zepaxid
BCP9000199
Diazepam-Eurogenerics
BIDD:PXR0158
SCHEMBL21442
Calmocitene
Servizepam
CCRIS 6009
Solis
7-Chloro-1,3-dihydro-1-methyl-5-phenyl-2H-1,4-benzodiazepin-2-one
Condition
T-quil
7-chloro-1-methyl-5-phenyl-1H-1,4-benzodiazepine-2(3H)on
DEA No. 2765
Tranqdyn
A3662/0155188
Freudal
Pacitran
HMS2051N04
Placidox 2
IDI1_000967
Psychopax
Kratium
Relaminal
Levium
MCULE-8990989144
Diazepan leo
Metil Gobanal
DivK1c_000967
NCGC00178168-03
DTXSID4020406
NINDS_000967
Elcion CR
NSC 169897
Tranquo-puren
AB02310
Diastat
Valeo
AN-23449
Diazemulus
Vanconin
Anxionil
Diazepam Elmu
Winii
Atilen
Diazepam Stada
Benzopin
S.A. R.L
Diazepamu
BRN 0754371
Seduksen
1-Methyl-5-phenyl-7-chloro-1,3-dihydro-2H-1,4-benzodiazepin-2-one
Calmpose
Sibazone
Ceregulart
Spectrum4_000576
7-Chloro-1-methyl-5-3H-1,4-benzodiazepin-2(1H)-one
D-Pam
Tox21_113071
7-Chloro-1-methyl-5-phenyl-2-oxo-2,3-dihydro-1H-benzo[f]-1,4-diazepine
Diacepan
Gewacalm
Parzam
Horizon
Plumiaz
Kabivitrum
Q3JTX2Q7TU
LA III
Renborin
Lovium
Metamidol
Dienpax
Mono (2-ethylhexyl) phthalate
Doval
Nellium
Dupin
Noan
Eurosan
NSC169897
Dialar
Umbrium
AKOS003367969
Diastat, Valium
Valium
Ansilive
Diazepam (JP17/USP/INN)
Vazepam
Apozepam
Diazepam Intensol
WY-3467
BCP0726000176
Diazepam, European Pharmacopoeia (EP) Reference Standard
Bialzepam
SAM001246536
C06948
Serenamin
2H-1, 7-chloro-1,3-dihydro-1-methyl-5-phenyl-
CC-26451
Sipam
7-chloranyl-1-methyl-5-phenyl-3H-1,4-benzodiazepin-2-one
CHEMBL12
Stesolin
7-chloro-1-methyl-5-phenyl-1,3-dihydro-[1,4]benzodiazepin-2-one
D0Y1YC
Tranimul
7-chloro-1-methyl-5-phenyl-3H-1,4-benzodiazepin-2-one
Ortopsique
GTPL3364
Paxum
I06-0194
Pro pam
KBioGR_001012
Quievita
Lamra
Ro-52807
Mandro-Zep
Diazepamum [INN-Latin]
Methyldiazepinone (pharmaceutical)
Dipezona
NCGC00178168-01
DSSTox_GSID_20406
neurolytr il
e-Pam
Novo-Dipam
Faustan
Diapine
Usempax ap
Alupram
Diazem
Valrelease
Antenex
Diazepam Dak
Vivol
Assival
Diazepam solution, 1.0 mg/mL in methanol, ampule of 1 mL, certified reference material
ZINC6427
BDBM50000766
Diazepam-Lipuro
BRD-K16508793-001-01-8
Sedapam
Calmociteno
Setonil
439-14-5
Centrazepam
Sonacon
7-chloro-1-methyl-1,3-dihydro-5-phenyl-2H-1,4-benzodiazepin-2-one
CPD000058398
Tensium
7-Chloro-1-methyl-5-phenyl-1H-benzo[e][1,4]diazepin-2(3H)-one
Desconet
Tranquase
A826456
Frustan
Paralium
HMS3393N04
Placidox 5
InChI=1/C16H13ClN2O/c1-19-14-8-7-12(17)9-13(14)16(18-10-15(19)20)11-5-3-2-4-6-11/h2-9H,10H2,1H
Q-pam
Kratium 2
Relanium
Liberetas
Medipam
Diazepin
MLS000759402
Dizac
NCGC00178168-04
Ducene
Nivalen
Eridan
NSC-169897
Tranquo-tablinen
AC-10561
Diastat (TN)
Valiquid
An-Ding
Vatran
Apaurin
Diazepam Fabra
WLN: T67 GNV JN IHJ CG G1 KR
Azedipamin
Diazepam [USAN:INN:BAN:JAN]
Best [Pharaceutical]
S.a. r.l.
Diazepamu [Polish]
BSPBio_003279
Seduxen
1-Methyl-5-phenyl-7-chloro-1,4-benzodiazepin-2-one
CAS-439-14-5
Sico Relax
53320-84-6
CHEBI:49575
Spectrum5_001890
7-Chloro-1-methyl-5-phenyl-1,3-dihydro-2H-1,4-benzodiazepin-2-one
D00293
Tox21_113071_1
7-Chloro-1-methyl-5-phenyl-2H-1,4-benzodiazepin-2-one
Diaceplex
NSC77518
Gihitan
Paxate
HSDB 3057
Pms-Diazepam
KBio1_000967
Quetinil
LA-111
Ro 5-2805
LS-122
Methyl diazepinone
Dipam
Morosan
Drenian
Nerozen
Duxen
Notense
Evacalm
Diapam
UNII-Q3JTX2Q7TU
Alboral
Diatran
Valium (TN)
Ansiolin
Diazepam 0.1 mg/ml in Methanol
Velium
Armonil
Diazepam Nordic
ZB000640
BCP23002
Diazepam, United States Pharmacopeia (USP) Reference Standard
BIDD:GT0105
Saromet
Calmaven
Serenzin
2H-1,4-Benzodiazepin-2-one, 7-chloro-1,3-dihydro-1-methyl-5-phenyl-
CCG-100997
SMR000058398
7-chloro-1,3-dihydro-1-methyl-5-phenyl-2H -1,4-benzodiazepin-2-one
Chuansuan
STK735517
7-Chloro-1-methyl-5-phenyl-1,4-benzodiazepin-2-one
DB00829
Trankinon
7-chloro-1-methyl-5-phenyl-3H-1,4-benzodiazepin-2-one.
Paceum
Gubex
Placidox 10
Iazepam
Prozepam
Kiatrium
Radizepam
Lembrol
Ruhsitus
Mandrozep
Diazepan
Methyldiazepinone, pharmaceutical
Disopam
NCGC00178168-02
DSSTox_RID_75566
Neurolytril
EINECS 207-122-5
Novodipam
Faustan,
AAOVKJBEBIDNHE-UHFFFAOYSA-N
Diaquel
Valaxona
Amiprol
Diazemuls
Valuzepam
Anxicalm
Diazepam Desitin
W-5055
Atensine
Diazepam solution, 1.0 mg/mL in methanol, analytical standard, for drug analysis
Zipan
Bensedin
Diazepam-ratiopharm
Britazepam
Sedipam
(Z)-7-chloro-1-methyl-5-phenyl-1H-benzo[e][1,4]diazepin-2(3H)-one
Calmod
Sibazon
5-24-04-00300 (Beilstein Handbook Reference)
Cercine
Spectrum3_001780
7-Chloro-1-methyl-2-oxo-5-phenyl-3H-1,4-benzodiazepine
CS-0653
Tensopam
7-chloro-1-methyl-5-phenyl-2,3-dihydro-1H-1,4-benzodiazepin-2-one
Desloneg
Tranquirit
FT-0632135
Paranten
HMS503A15
Plidan
Jinpanfan
Q-Pam Relanium
LA 111
Reliver
Lizan
Mentalium
Diazetard
MLS001424086
Domalium
NCGC00260007-01
Duksen
Nixtensyn
Euphorin P
NSC-77518 [ Show all ] |
|---|


![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417