

You can:
| Name | Prostaglandin E2 receptor EP4 subtype |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | PTGER4 |
| Synonym | Prostanoid EP4 receptor PGE2 receptor EP4 subtype PGE receptor EP4 subtype EP4 receptor EP2 |
| Disease | Ulcerative colitis Glaucoma Inflammatory disease Migraine Osteoarthritis [ Show all ] |
| Length | 488 |
| Amino acid sequence | MSTPGVNSSASLSPDRLNSPVTIPAVMFIFGVVGNLVAIVVLCKSRKEQKETTFYTLVCGLAVTDLLGTLLVSPVTIATYMKGQWPGGQPLCEYSTFILLFFSLSGLSIICAMSVERYLAINHAYFYSHYVDKRLAGLTLFAVYASNVLFCALPNMGLGSSRLQYPDTWCFIDWTTNVTAHAAYSYMYAGFSSFLILATVLCNVLVCGALLRMHRQFMRRTSLGTEQHHAAAAASVASRGHPAASPALPRLSDFRRRRSFRRIAGAEIQMVILLIATSLVVLICSIPLVVRVFVNQLYQPSLEREVSKNPDLQAIRIASVNPILDPWIYILLRKTVLSKAIEKIKCLFCRIGGSRRERSGQHCSDSQRTSSAMSGHSRSFISRELKEISSTSQTLLPDLSLPDLSENGLGGRNLLPGVPGMGLAQEDTTSLRTLRISETSDSSQGQDSESVLLVDEAGGSGRAGPAPKGSSLQVTFPSETLNLSEKCI |
| UniProt | P35408 |
| Protein Data Bank | 5ywy, 5yhl |
| GPCR-HGmod model | P35408 |
| 3D structure model | This structure is from PDB ID 5ywy. |
| BioLiP | BL0434347, BL0434289 |
| Therapeutic Target Database | T18876 |
| ChEMBL | CHEMBL1836 |
| IUPHAR | 343 |
| DrugBank | BE0003522 |
| Name | Prostaglandin E2 |
|---|---|
| Molecular formula | C20H32O5 |
| IUPAC name | (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxocyclopentyl]hept-5-enoic acid |
| Molecular weight | 352.471 |
| Hydrogen bond acceptor | 5 |
| Hydrogen bond donor | 3 |
| XlogP | 2.8 |
| Synonyms | Dinoprostone, United States Pharmacopeia (USP) Reference Standard (Z)-7-[(1r,2r,3r)-3-Hydroxy-2-[(E,3s)-3-Hydroxyoct-1-Enyl]-5-Oxo-Cyclopentyl]hept-5-Enoic Acid E2, Prostaglandin 1798-EP2301922A1 HMS1361K12 [ Show all ] |
| Inchi Key | XEYBRNLFEZDVAW-ARSRFYASSA-N |
| Inchi ID | InChI=1S/C20H32O5/c1-2-3-6-9-15(21)12-13-17-16(18(22)14-19(17)23)10-7-4-5-8-11-20(24)25/h4,7,12-13,15-17,19,21,23H,2-3,5-6,8-11,14H2,1H3,(H,24,25)/b7-4-,13-12+/t15-,16+,17+,19+/m0/s1 |
| PubChem CID | 5280360 |
| ChEMBL | CHEMBL548 |
| IUPHAR | 1883, 1916 |
| BindingDB | 35847 |
| DrugBank | DB00917 |
Structure | 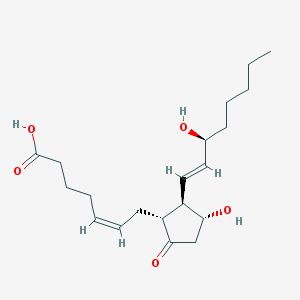 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| N/A | N/A | DrugBank | |
| Activity | 40.0 % | PMID24279689 | ChEMBL |
| EC50 | 3.0 nM | PMID12643927, PMID18039575 | BindingDB,ChEMBL |
| EC50 | 7.5 nM | PMID26985320 | BindingDB,ChEMBL |
| EC50 | 900.0 nM | PMID19584306 | IUPHAR |
| IC50 | 0.45 nM | PMID23466604 | ChEMBL |
| IC50 | 0.55 nM | PMID27876250 | ChEMBL |
| IC50 | 0.7 nM | PMID12643927 | BindingDB |
| IC50 | 0.7 nM | PMID12643927 | ChEMBL |
| IC50 | 1.1 nM | PMID23403082 | ChEMBL |
| IC50 | 3.8 nM | PMID18983139 | ChEMBL |
| IC50 | 5.5 nM | PMID27876250 | BindingDB |
| IC50 | 9.9 nM | PMID20218623 | BindingDB,ChEMBL |
| Inhibition | 5.0 % | PMID23403082 | ChEMBL |
| Kd | 0.3 - 24.0 nM | PMID16604093, PMID10634944, PMID10952683, PMID22480736 | IUPHAR |
| Ki | 0.17 nM | PMID23466604 | ChEMBL |
| Ki | 0.45 nM | PMID23403082 | ChEMBL |
| Ki | 0.79 nM | PMID18039575, PMID17931866, PMID10634944, PMID17531488 | BindingDB,ChEMBL |
| Ki | 0.794 - 7.94 nM | PMID16604093, PMID10634944, PMID10462542, PMID10952683, PMID17495127 | IUPHAR |
| Ki | 1.9 nM | PMID18983139 | ChEMBL |
| Ki | 3.1 nM | PMID24279689 | BindingDB,ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417