| Synonyms | SPBio_002188
Tox21_110217
WYWIFABBXFUGLM-UHFFFAOYSA-N
Operil (TN)
CC-33432
Oxymetazoline (INN)
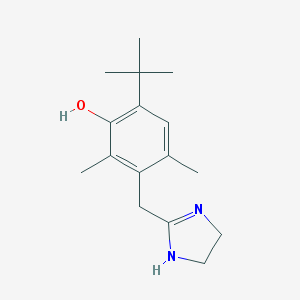
Phenol, 3-[(4,5-dihydro-1H-imidazol-2-yl)methyl]-6-(1,1-dimethylethyl)-2,4-dimethyl-
DSSTox_GSID_40691
Prestwick0_000224
Genasal Nasal Spray Up to 12 Hour Relief
Iliadin
6-t-Butyl-3-(2-imidazolin-2-ylmethyl)-2,4-dimethylphenol
KBioSS_001531
8VLN5B44ZY
Nasivine (Salt/Mix)
Afrin Cherry 12 Hour Nasal Spray
NCGC00015766-06
AK113123
NCGC00022345-02
BRD-K16195444-003-16-1
SBI-0050878.P004
Spectrum4_000464
Vicks Sinex
BSPBio_002145
Oximetazolinum
Oxymetazolinum
DivK1c_000567
Phenol, 6-t-butyl-3-(2-imidazolin-2-ylmethyl)-2,4-dimethyl-
Duramist Plus Up To 12 Hour Nasal Decongestant Spray
Prestwick3_000224
H-990
1491-59-4
KBio2_001531
6-tert-Butyl-3-(2-imidazolin-2-ylmethyl)-2,4-dimethylphenol
Lopac0_000903
AB00053513_14
NCGC00015766-01
Afrin Original 12 Hour Nose Drops
NCGC00015766-09
BDBM30712
NCGC00022345-06
Sinerol
STK075254
Visine L.R
NINDS_000567
CAS-1491-59-4
oxymetazolina
D08322
Oxymetozoline
Drixoral Nasal Solution
Phenol, 6-tert-butyl-3-(2-imidazolin-2-ylmethyl)-2,4-dimethyl-(7CI,8CI)
FT-0601532
HSDB 3143
3-[(4,5-Dihydro-1H-imidazol-2-yl)methyl]-6-(1,1-dimethylethyl)-2,4-dimethylphenol
KBio3_001645
6-tert-Butyl-3-(4,5-dihydro-1H-imidazol-2-ylmethyl)-2,4-dimethylphenol
Nafrine
AC1L1IM1
NCGC00015766-04
Afrin, Dristan12-hrnasalspray, Ocuclear, Drixine
NCGC00015766-13
BPBio1_000419
Rhinofrenol
Spectrum2_000998
Tox21_110217_1
ZINC57435
Oximetazolina
CCG-204985
oxymetazoline hydrochloride crystalline
DB00935
Phenol, 3-[(4,5-dihydro-1H-imidazol-2-yl)methyl]-6-(1,1-dimethylethyl)-2,4-dimethyl- (9CI)
DSSTox_RID_79545
Prestwick1_000224
GTPL124
KB-302508
6-tert-butyl-3-((4,5-dihydro-1H-imidazol-2-yl)methyl)-2,4-dimethylphenol
L000459
AB00053513-12
Navasin
Afrin Extra Moisturizing 12 Hour Nasal Spray
NCGC00015766-07
AKOS007930348
NCGC00022345-04
BRN 0886303
SCH-9384
Spectrum5_001114
Vicks Sinex 12 Hour Nasal Spray
C-16261
Oxylazine
CHEMBL762
Oxymetazolinum [INN-Latin]
Dristan Long Lasting Mentholated Nasal Spray
Phenol, 6-tert-butyl-3-(2-imidazolin-2-ylmethyl)-2,4-dimethyl-
EINECS 216-079-1
Hazol
2-(4-tert-Butyl-2,6-dimethyl-3-hydroxybenzyl)-2-imidazoline
KBio2_004099
6-tert-Butyl-3-(4,5-dihydro-1H-imidazol-2-ylmethyl)-2,4-dimethyl-phenol
LS-104146
AB00053513_15
NCGC00015766-02
Afrin Original 12 Hour Pump Mist
NCGC00015766-10
Biomol-NT_000161
Neo-Synephrine 12 Hour Spray
SPBio_001095
Tocris-1142
Visine L.R.
Nostrilla 12 Hour Nasal Decongestant
CAS-151615
D09EBS
Phenol, 3-((4,5-dihydro-1H-imidazol-2-yl)methyl)-6-(1,1-dimethylethyl)-2,4-dimethyl-
DSSTox_CID_20691
Phenol,3-[(4,5-dihydro-1H-imidazol-2-yl)methyl]-6-(1,1-dimethylethyl)-2,4-dimethyl-
FT-0650596
IDI1_000567
KBioGR_000908
6-tert-Butyl-3-(4,5-dihydro-1H-imidazol-2-ylmethyl)-2,4-dimethylphenol #
Nasal Relief 12 Hour Nasal Spray
Afrin
NCGC00015766-05
AJ-09693
NCGC00015766-16
BRD-K16195444-001-01-7
Rhinolitan
Spectrum3_000533
UNII-8VLN5B44ZY
Oximetazolina [INN-Spanish]
CHEBI:7862
Oxymetazoline [INN:BAN]
Decongestant (TN)
Phenol, 3-[(4,5-dihydro-1H-imidazol-2-yl)methyl]-6-(1,1-dimethylethyl)-2,4-dimethyl-(9CI)
DTXSID3040691
Prestwick2_000224
H 990
KBio1_000567
6-tert-butyl-3-(2-imidazolin-2-ylmethyl)-2,4-dimethyl-phenol;hydrochloride
Lopac-O-2378
AB00053513-13
Navisin
Afrin Original 12 Hour Nasal Spray
NCGC00015766-08
AN-10265
NCGC00022345-05
BSPBio_000267
SCHEMBL24301
Spectrum_001051
Vicks Sinex 12 Hour Ultra Fine Mist for Sinus Relief
C07363
oxymeta zoline
cid_66259
Oxymethazoline
Dristan Long Lasting Nasal Mist
Phenol, 6-tert-butyl-3-(2-imidazolin-2-ylmethyl)-2,4-dimethyl- (7CI,8CI)
FT-0082560
HMS2089G03
3-(4,5-dihydro-1H-imidazol-2-ylmethyl)-2,4-dimethyl-6-tert-butyl-phenol
KBio2_006667
6-tert-butyl-3-(4,5-dihydro-1H-imidazol-2-ylmethyl)-2,4-dimethyl-phenol;hydrochloride
MCULE-8690625159
AC-6370
NCGC00015766-03
Afrin Sinus 12 Hour Nasal Spray
NCGC00015766-11
BPBio1_000295
Nezeril [ Show all ] |
|---|


![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417