

You can:
| Name | 5-hydroxytryptamine receptor 1A |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | HTR1A |
| Synonym | 5-HT-1A 5-HT1A serotonin receptor 1A 5-HT1A receptor 5-hydroxytryptamine (serotonin) receptor 1A, G protein-coupled [ Show all ] |
| Disease | Urinary incontinence Generalized anxiety disorder Generalized anxiety disorder; Social phobia Hypertension Hypoactive sexual desire disorder [ Show all ] |
| Length | 422 |
| Amino acid sequence | MDVLSPGQGNNTTSPPAPFETGGNTTGISDVTVSYQVITSLLLGTLIFCAVLGNACVVAAIALERSLQNVANYLIGSLAVTDLMVSVLVLPMAALYQVLNKWTLGQVTCDLFIALDVLCCTSSILHLCAIALDRYWAITDPIDYVNKRTPRRAAALISLTWLIGFLISIPPMLGWRTPEDRSDPDACTISKDHGYTIYSTFGAFYIPLLLMLVLYGRIFRAARFRIRKTVKKVEKTGADTRHGASPAPQPKKSVNGESGSRNWRLGVESKAGGALCANGAVRQGDDGAALEVIEVHRVGNSKEHLPLPSEAGPTPCAPASFERKNERNAEAKRKMALARERKTVKTLGIIMGTFILCWLPFFIVALVLPFCESSCHMPTLLGAIINWLGYSNSLLNPVIYAYFNKDFQNAFKKIIKCKFCRQ |
| UniProt | P08908 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | P08908 |
| 3D structure model | This predicted structure model is from GPCR-EXP P08908. |
| BioLiP | N/A |
| Therapeutic Target Database | T78709 |
| ChEMBL | CHEMBL214 |
| IUPHAR | 1 |
| DrugBank | BE0000291 |
| Name | 5-Carboxamidotryptamine |
|---|---|
| Molecular formula | C11H13N3O |
| IUPAC name | 3-(2-aminoethyl)-1H-indole-5-carboxamide |
| Molecular weight | 203.245 |
| Hydrogen bond acceptor | 2 |
| Hydrogen bond donor | 3 |
| XlogP | -0.6 |
| Synonyms | 5-CT Biomol-NT_000109 D02UWB NCGC00015182-02 PDSP2_000762 [ Show all ] |
| Inchi Key | WKZLNEWVIAGNAW-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C11H13N3O/c12-4-3-8-6-14-10-2-1-7(11(13)15)5-9(8)10/h1-2,5-6,14H,3-4,12H2,(H2,13,15) |
| PubChem CID | 1809 |
| ChEMBL | CHEMBL18840 |
| IUPHAR | 4 |
| BindingDB | 21392 |
| DrugBank | N/A |
Structure | 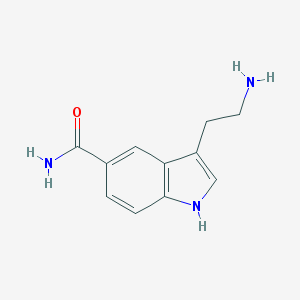 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| %max | 96.0 - | PMID18817363 | ChEMBL |
| IC50 | 0.2951 nM | Bioorg. Med. Chem. Lett., (1994) 4:10:1207 | ChEMBL |
| IC50 | 0.3 nM | N/A | BindingDB |
| Kd | 0.3981 nM | PMID19754201 | ChEMBL |
| Ki | 0.0501187 - 0.398107 nM | PMID1386736, PMID9760039, PMID9550290, PMID9205951 | IUPHAR |
| Ki | 0.2 nM | PMID2078271 | BindingDB |
| Ki | >0.3 nM | PMID10753471 | ChEMBL |
| Ki | 0.34 nM | PMID7984267 | PDSP,BindingDB |
| Ki | 0.39 nM | Hoyer et al., PMID1986 | PDSP |
| Ki | 0.4 nM | Hoyer et al., PMID1986 | PDSP |
| Ki | 0.52 nM | PMID7984267 | PDSP,BindingDB |
| Ki | 0.53 nM | PMID8461029 | PDSP |
| Ki | 0.631 nM | PMID14613313 | ChEMBL |
| Ki | 0.85 nM | PMID7984267 | PDSP |
| Ki | 17.37 nM | PMID7984267 | PDSP,BindingDB |
| Ki | 17.5 nM | PMID8461029 | PDSP,BindingDB |
| Max | 96.0 % | PMID10514291, PMID10425105 | ChEMBL |
| pD2 | 8.45 - | PMID10514291, PMID10425105, PMID18817363 | ChEMBL |
| pKD | 9.5 - | PMID8515429 | ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417