

You can:
| Name | D(2) dopamine receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | DRD2 |
| Synonym | dopamine receptor 2 Dopamine D2 receptor D2R D2A and D2B D2(415) and D2(444) [ Show all ] |
| Disease | Substance dependence Major depressive disorder Maintain blood pressure in hypotensive states Insomnia Inflammatory disease [ Show all ] |
| Length | 443 |
| Amino acid sequence | MDPLNLSWYDDDLERQNWSRPFNGSDGKADRPHYNYYATLLTLLIAVIVFGNVLVCMAVSREKALQTTTNYLIVSLAVADLLVATLVMPWVVYLEVVGEWKFSRIHCDIFVTLDVMMCTASILNLCAISIDRYTAVAMPMLYNTRYSSKRRVTVMISIVWVLSFTISCPLLFGLNNADQNECIIANPAFVVYSSIVSFYVPFIVTLLVYIKIYIVLRRRRKRVNTKRSSRAFRAHLRAPLKGNCTHPEDMKLCTVIMKSNGSFPVNRRRVEAARRAQELEMEMLSSTSPPERTRYSPIPPSHHQLTLPDPSHHGLHSTPDSPAKPEKNGHAKDHPKIAKIFEIQTMPNGKTRTSLKTMSRRKLSQQKEKKATQMLAIVLGVFIICWLPFFITHILNIHCDCNIPPVLYSAFTWLGYVNSAVNPIIYTTFNIEFRKAFLKILHC |
| UniProt | P14416 |
| Protein Data Bank | 6cm4, 6c38 |
| GPCR-HGmod model | P14416 |
| 3D structure model | This structure is from PDB ID 6cm4. |
| BioLiP | BL0408379, BL0403379 |
| Therapeutic Target Database | T67162 |
| ChEMBL | CHEMBL217 |
| IUPHAR | 215 |
| DrugBank | BE0000756 |
| Name | RACLOPRIDE |
|---|---|
| Molecular formula | C15H20Cl2N2O3 |
| IUPAC name | 3,5-dichloro-N-[[(2S)-1-ethylpyrrolidin-2-yl]methyl]-2-hydroxy-6-methoxybenzamide |
| Molecular weight | 347.236 |
| Hydrogen bond acceptor | 4 |
| Hydrogen bond donor | 2 |
| XlogP | 2.9 |
| Synonyms | DB12518 HMS2051C03 MLS000758220 NCGC00025303-05 Raclopride;3,5-Dichloro-N-(1-ethyl-pyrrolidin-2-ylmethyl)-2-hydroxy-6-methoxy-benzamide [ Show all ] |
| Inchi Key | WAOQONBSWFLFPE-VIFPVBQESA-N |
| Inchi ID | InChI=1S/C15H20Cl2N2O3/c1-3-19-6-4-5-9(19)8-18-15(21)12-13(20)10(16)7-11(17)14(12)22-2/h7,9,20H,3-6,8H2,1-2H3,(H,18,21)/t9-/m0/s1 |
| PubChem CID | 3033769 |
| ChEMBL | CHEMBL8809 |
| IUPHAR | 3299, 94 |
| BindingDB | 50005118 |
| DrugBank | DB12518 |
Structure | 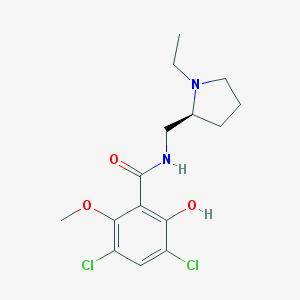 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| N/A | N/A | DrugBank | |
| Ki | 0.6 nM | PMID8099194 | BindingDB |
| Ki | 0.64 nM | PMID9577836, PMID9015795 | BindingDB |
| Ki | 1.2 nM | PMID24779610 | BindingDB |
| Ki | 1.21 nM | PMID24779610 | ChEMBL |
| Ki | 2.0 nM | PMID8102973 | BindingDB |
| Ki | 3.2 nM | PMID1840645 | BindingDB |
| Ki | 10.0 nM | PMID7473180 | IUPHAR |
| Ki | 10.5 nM | PMID1840645 | BindingDB |
| Ki | 12.7 nM | PMID24848155 | ChEMBL |
| Ki | 13.0 nM | PMID24848155 | BindingDB |
| Ki | 17.0 nM | PMID17765546 | BindingDB,ChEMBL |
| Ki | 31.0 nM | PMID17765546 | BindingDB,ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417