

You can:
| Name | Alpha-1A adrenergic receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | ADRA1A |
| Synonym | alpha1a ADRA1C ADRA1L1 adrenergic alpha 1c receptor adrenergic receptor alpha 1c [ Show all ] |
| Disease | Urinary incontinence Benign prostatic hyperplasia Cognitive disorders Female sexual dysfunction Glaucoma [ Show all ] |
| Length | 466 |
| Amino acid sequence | MVFLSGNASDSSNCTQPPAPVNISKAILLGVILGGLILFGVLGNILVILSVACHRHLHSVTHYYIVNLAVADLLLTSTVLPFSAIFEVLGYWAFGRVFCNIWAAVDVLCCTASIMGLCIISIDRYIGVSYPLRYPTIVTQRRGLMALLCVWALSLVISIGPLFGWRQPAPEDETICQINEEPGYVLFSALGSFYLPLAIILVMYCRVYVVAKRESRGLKSGLKTDKSDSEQVTLRIHRKNAPAGGSGMASAKTKTHFSVRLLKFSREKKAAKTLGIVVGCFVLCWLPFFLVMPIGSFFPDFKPSETVFKIVFWLGYLNSCINPIIYPCSSQEFKKAFQNVLRIQCLCRKQSSKHALGYTLHPPSQAVEGQHKDMVRIPVGSRETFYRISKTDGVCEWKFFSSMPRGSARITVSKDQSSCTTARVRSKSFLQVCCCVGPSTPSLDKNHQVPTIKVHTISLSENGEEV |
| UniProt | P35348 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | P35348 |
| 3D structure model | This predicted structure model is from GPCR-EXP P35348. |
| BioLiP | N/A |
| Therapeutic Target Database | T92609 |
| ChEMBL | CHEMBL229 |
| IUPHAR | 22 |
| DrugBank | BE0000501 |
| Name | terazosin |
|---|---|
| Molecular formula | C19H25N5O4 |
| IUPAC name | [4-(4-amino-6,7-dimethoxyquinazolin-2-yl)piperazin-1-yl]-(oxolan-2-yl)methanone |
| Molecular weight | 387.44 |
| Hydrogen bond acceptor | 8 |
| Hydrogen bond donor | 1 |
| XlogP | 1.4 |
| Synonyms | GTPL7302 Hytracin MET029 NCGC00016026-04 1-(4-amino-6,7-dimethoxy-2-quinazolinyl)4-[(tetrahydro-2-furanyl)carbonyl]piperazine [ Show all ] |
| Inchi Key | VCKUSRYTPJJLNI-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C19H25N5O4/c1-26-15-10-12-13(11-16(15)27-2)21-19(22-17(12)20)24-7-5-23(6-8-24)18(25)14-4-3-9-28-14/h10-11,14H,3-9H2,1-2H3,(H2,20,21,22) |
| PubChem CID | 5401 |
| ChEMBL | CHEMBL611 |
| IUPHAR | 7302 |
| BindingDB | 50033111 |
| DrugBank | DB01162 |
Structure | 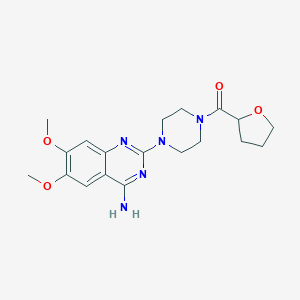 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| N/A | N/A | DrugBank | |
| EC50 | 51.0 nM | PMID25813897 | BindingDB,ChEMBL |
| EC50 | 51.89 nM | PMID27658792 | ChEMBL |
| EC50 | 52.0 nM | PMID27658792 | BindingDB |
| Kd | 36.0 nM | PMID9379432 | BindingDB |
| Kd | 36.31 nM | PMID9379432, PMID11384242 | ChEMBL |
| Ki | 1.8197 nM | PMID11805207 | PDSP |
| Ki | 1.82 nM | PMID11805207 | BindingDB |
| Ki | 2.0 nM | PMID9873563, PMID9379432 | BindingDB,IUPHAR,ChEMBL |
| Ki | 2.4 nM | PMID9873563 | BindingDB,ChEMBL |
| Ki | 2.5 nM | PMID16420037 | BindingDB |
| Ki | 2.9 nM | PMID9873563 | BindingDB,ChEMBL |
| Ki | 3.98 nM | PMID7815325 | BindingDB |
| Ki | 4.0 nM | PMID10579840 | BindingDB,ChEMBL |
| Ki | 4.2 nM | PMID9873563, PMID9548811 | BindingDB,ChEMBL |
| Ki | 6.3 nM | PMID16420037 | BindingDB |
| Ki | 6.9 nM | PMID10579840, PMID7752182, PMID10579842, PMID10579841 | BindingDB,ChEMBL |
| Ki | 6.918 nM | PMID9651170 | ChEMBL |
| Ki | 7.3 nM | PMID7658428 | BindingDB,ChEMBL |
| Ki | 25.11 nM | PMID7815325 | BindingDB |
| Ki | 32.0 nM | PMID9873563 | BindingDB,ChEMBL |
| Ki | 39.81 nM | PMID9135028 | ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417