

You can:
| Name | Somatostatin receptor type 1 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | SSTR1 |
| Synonym | SRIF-2 SS-1-R SS1-R SS1R SST1 receptor |
| Disease | Alzheimer disease Cushing's disease Neuroendocrine cancer |
| Length | 391 |
| Amino acid sequence | MFPNGTASSPSSSPSPSPGSCGEGGGSRGPGAGAADGMEEPGRNASQNGTLSEGQGSAILISFIYSVVCLVGLCGNSMVIYVILRYAKMKTATNIYILNLAIADELLMLSVPFLVTSTLLRHWPFGALLCRLVLSVDAVNMFTSIYCLTVLSVDRYVAVVHPIKAARYRRPTVAKVVNLGVWVLSLLVILPIVVFSRTAANSDGTVACNMLMPEPAQRWLVGFVLYTFLMGFLLPVGAICLCYVLIIAKMRMVALKAGWQQRKRSERKITLMVMMVVMVFVICWMPFYVVQLVNVFAEQDDATVSQLSVILGYANSCANPILYGFLSDNFKRSFQRILCLSWMDNAAEEPVDYYATALKSRAYSVEDFQPENLESGGVFRNGTCTSRITTL |
| UniProt | P30872 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | P30872 |
| 3D structure model | This predicted structure model is from GPCR-EXP P30872. |
| BioLiP | N/A |
| Therapeutic Target Database | T16633 |
| ChEMBL | CHEMBL1917 |
| IUPHAR | 355 |
| DrugBank | BE0000452 |
| Name | BIM-23027 |
|---|---|
| Molecular formula | C43H54N8O7 |
| IUPAC name | (3S,6S,9S,12R,15S,18S)-9-(4-aminobutyl)-3-benzyl-6-ethyl-15-[(4-hydroxyphenyl)methyl]-12-(1H-indol-3-ylmethyl)-1,18-dimethyl-1,4,7,10,13,16-hexazacyclooctadecane-2,5,8,11,14,17-hexone |
| Molecular weight | 794.954 |
| Hydrogen bond acceptor | 8 |
| Hydrogen bond donor | 8 |
| XlogP | 3.5 |
| Synonyms | BIM 23027 N-Methylcyclo[L-Ala-L-Tyr-D-Trp-L-Lys-L-Abu-L-Phe-] BDBM82455 cyclo(N-Me-Ala-Tyr-d-Trp-Lys-Abu-Phe) |
| Inchi Key | SKOFEUKLEYXDDV-GXTZDVISSA-N |
| Inchi ID | InChI=1S/C43H54N8O7/c1-4-32-39(54)50-37(23-27-12-6-5-7-13-27)43(58)51(3)26(2)38(53)48-35(22-28-17-19-30(52)20-18-28)41(56)49-36(24-29-25-45-33-15-9-8-14-31(29)33)42(57)47-34(40(55)46-32)16-10-11-21-44/h5-9,12-15,17-20,25-26,32,34-37,45,52H,4,10-11,16,21-24,44H2,1-3H3,(H,46,55)(H,47,57)(H,48,53)(H,49,56)(H,50,54)/t26-,32-,34-,35-,36+,37-/m0/s1 |
| PubChem CID | 9940596 |
| ChEMBL | N/A |
| IUPHAR | N/A |
| BindingDB | 82455 |
| DrugBank | N/A |
Structure | 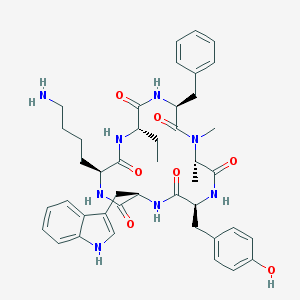 |
| Lipinski's druglikeness | This ligand has more than 5 hydrogen bond donor. This ligand is heavier than 500 daltons. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| Ki | 1000.0 nM | PMID8100350, PMID7870182 | BindingDB |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417