

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 | |
| | | | | | | | | | | | | | | | | | | | | |
| MFPNGTASSPSSSPSPSPGSCGEGGGSRGPGAGAADGMEEPGRNASQNGTLSEGQGSAILISFIYSVVCLVGLCGNSMVIYVILRYAKMKTATNIYILNLAIADELLMLSVPFLVTSTLLRHWPFGALLCRLVLSVDAVNMFTSIYCLTVLSVDRYVAVVHPIKAARYRRPTVAKVVNLGVWVLSLLVILPIVVFSRTAANSDGTVACNMLMPEPAQRWLVGFVLYTFLMGFLLPVGAICLCYVLIIAKMRMVALKAGWQQRKRSERKITLMVMMVVMVFVICWMPFYVVQLVNVFAEQDDATVSQLSVILGYANSCANPILYGFLSDNFKRSFQRILCLSWMDNAAEEPVDYYATALKSRAYSVEDFQPENLESGGVFRNGTCTSRITTL | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCSSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHCCCSSSSSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 9888888888788999986556778988998887656678888777666656522789859999999999998778863022010887787999999999999999999689999999858998856788899999999999999999999997387754006543243678899988999999999997999871558878997899731895688999999999999999999999999999999998276788752032212423677507899999999869999999999875314699999999999998698999998099999999998444316875567877677666567754677888878788875699777578779 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 | |
| | | | | | | | | | | | | | | | | | | | | |
| MFPNGTASSPSSSPSPSPGSCGEGGGSRGPGAGAADGMEEPGRNASQNGTLSEGQGSAILISFIYSVVCLVGLCGNSMVIYVILRYAKMKTATNIYILNLAIADELLMLSVPFLVTSTLLRHWPFGALLCRLVLSVDAVNMFTSIYCLTVLSVDRYVAVVHPIKAARYRRPTVAKVVNLGVWVLSLLVILPIVVFSRTAANSDGTVACNMLMPEPAQRWLVGFVLYTFLMGFLLPVGAICLCYVLIIAKMRMVALKAGWQQRKRSERKITLMVMMVVMVFVICWMPFYVVQLVNVFAEQDDATVSQLSVILGYANSCANPILYGFLSDNFKRSFQRILCLSWMDNAAEEPVDYYATALKSRAYSVEDFQPENLESGGVFRNGTCTSRITTL | |
| 7434433233232231332313343333323332222233333332433424333000000022012002202311200000001033210000000000010001000000000001026201002001100100131001000000000020000000000202321321000000000110000001000000202437522100101014433302000101101212231120002000100110233344443444434310000000000000000021000000000014332100100000000200020000000004401410140021214443454435344343443344345344443444433444334334136 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 | | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCSSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHCCCSSSSSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MFPNGTASSPSSSPSPSPGSCGEGGGSRGPGAGAADGMEEPGRNASQNGTLSEGQGSAILISFIYSVVCLVGLCGNSMVIYVILRYAKMKTATNIYILNLAIADELLMLSVPFLVTSTLLRHWPFGALLCRLVLSVDAVNMFTSIYCLTVLSVDRYVAVVHPIKAARYRRPTVAKVVNLGVWVLSLLVILPIVVFSRTAANSDGTVACNMLMPEPAQRWLVGFVLYTFLMGFLLPVGAICLCYVLIIAKMRMVALKAGWQQRKRSERKITLMVMMVVMVFVICWMPFYVVQLVNVFAEQDDATVSQLSVILGYANSCANPILYGFLSDNFKRSFQRILCLSWMDNAAEEPVDYYATALKSRAYSVEDFQPENLESGGVFRNGTCTSRITTL | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.44 | 0.35 | 0.76 | 3.20 | Download | ---------------------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPR-DGAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------------------------ | |||||||||||||||||||
| 2 | 4n6hA | 0.45 | 0.36 | 0.75 | 3.97 | Download | --------------------------------------------GSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPR-DGAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLC----------------------------------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.43 | 0.35 | 0.76 | 4.02 | Download | ---------------------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDG-AVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------------------------ | |||||||||||||||||||
| 4 | 4djh | 0.43 | 0.33 | 0.73 | 1.59 | Download | -----------------------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRDVDVIECSLQFPDDDYSWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA-ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP--------------------------------------------------- | |||||||||||||||||||
| 5 | 4djh | 0.43 | 0.33 | 0.73 | 1.18 | Download | -----------------------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRDVDVIECSLQFPDDDYWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA-ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP--------------------------------------------------- | |||||||||||||||||||
| 6 | 4n6hA | 0.44 | 0.35 | 0.75 | 3.37 | Download | -----------------------------------------------GARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPR-DGAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------------------------ | |||||||||||||||||||
| 7 | 4djh | 0.43 | 0.33 | 0.72 | 1.74 | Download | -------------------------------------------------------AIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRDVDVIECSLQFPDDDYSWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA-ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCF---------------------------------------------------- | |||||||||||||||||||
| 8 | 4djhA | 0.43 | 0.33 | 0.73 | 5.37 | Download | -----------------------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDDVIECSLQFPDDYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALG-SAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP--------------------------------------------------- | |||||||||||||||||||
| 9 | 4n6hA | 0.44 | 0.35 | 0.76 | 3.37 | Download | ---------------------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPR-DGAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------------------------ | |||||||||||||||||||
| 10 | 5c1mA | 0.43 | 0.33 | 0.74 | 3.80 | Download | ------------------------------------------GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVRYTKMKTATNIYIFNLALADALATSTLPFQSVNYLMGTWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRTPRNAKIVNVCNWILSSAIGLPVMFMATTKYR-QGSIDCTLTFSHPTWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMLSGSKEKDRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKALITTFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF--------------------------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
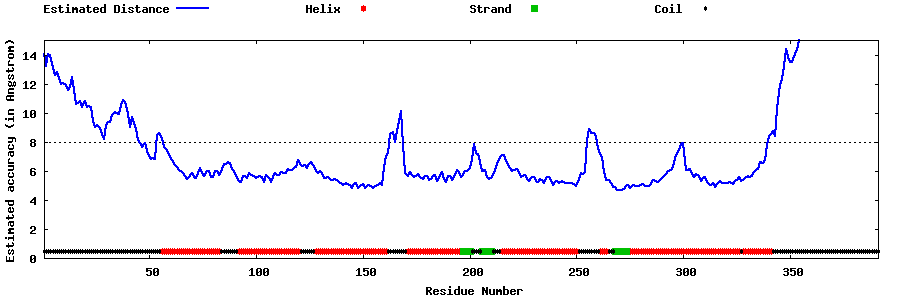
| Generated 3D models | Estimated local accuracy of models | ||
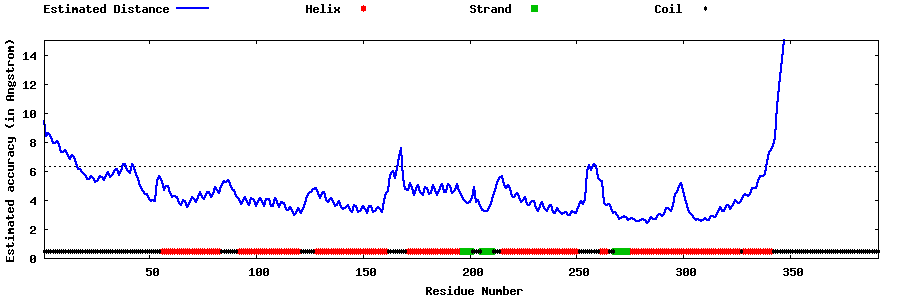 | |||
|
|
|||
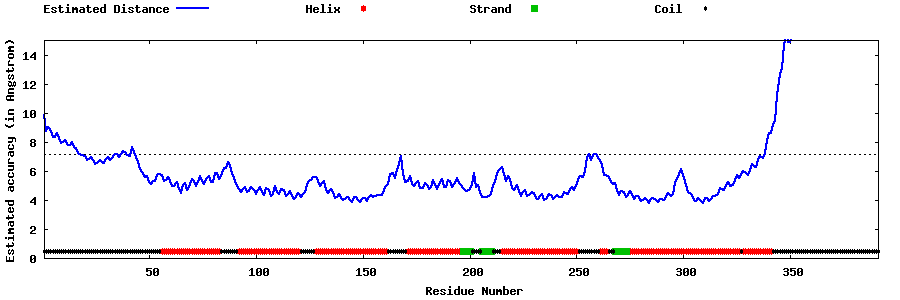 | |||
|
| |||
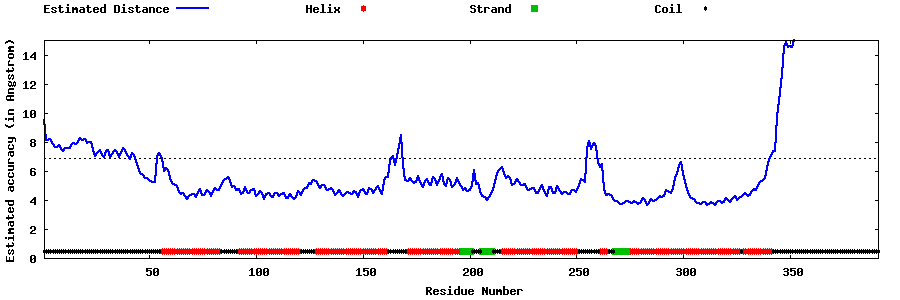 | |||
|
| |||
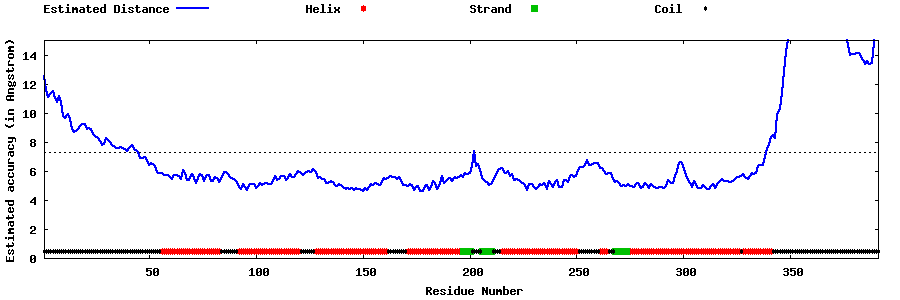 | |||
|
| |||
 | |||
|
| |||