

You can:
| Name | Muscarinic acetylcholine receptor M2 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | CHRM2 |
| Synonym | cholinergic receptor AChR M2 M2 muscarinic acetylcholine receptor M2 receptor Chrm-2 [ Show all ] |
| Disease | Urinary incontinence Heart failure Nausea; Addiction Parkinson's disease Peptic ulcer [ Show all ] |
| Length | 466 |
| Amino acid sequence | MNNSTNSSNNSLALTSPYKTFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIVGVRTVEDGECYIQFFSNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRIKKDKKEPVANQDPVSPSLVQGRIVKPNNNNMPSSDDGLEHNKIQNGKAPRDPVTENCVQGEEKESSNDSTSVSAVASNMRDDEITQDENTVSTSLGHSKDENSKQTCIRIGTKTPKSDSCTPTNTTVEVVGSSGQNGDEKQNIVARKIVKMTKQPAKKKPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPCIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLMCHYKNIGATR |
| UniProt | P08172 |
| Protein Data Bank | 5zkc, 4mqs, 4mqt, 5yc8, 5zk3, 5zkb, 5zk8 |
| GPCR-HGmod model | P08172 |
| 3D structure model | This structure is from PDB ID 5zkc. |
| BioLiP | BL0433341, BL0433216, BL0263147, BL0263146, BL0263145, BL0433339, BL0433340, BL0433338 |
| Therapeutic Target Database | T46185 |
| ChEMBL | CHEMBL211 |
| IUPHAR | 14 |
| DrugBank | BE0000560 |
| Name | pirenzepine |
|---|---|
| Molecular formula | C19H21N5O2 |
| IUPAC name | 11-[2-(4-methylpiperazin-1-yl)acetyl]-5H-pyrido[2,3-b][1,4]benzodiazepin-6-one |
| Molecular weight | 351.41 |
| Hydrogen bond acceptor | 5 |
| Hydrogen bond donor | 1 |
| XlogP | 0.1 |
| Synonyms | Spectrum_001378 FT-0600051 KBio1_000127 L000485 NCGC00015836-03 [ Show all ] |
| Inchi Key | RMHMFHUVIITRHF-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C19H21N5O2/c1-22-9-11-23(12-10-22)13-17(25)24-16-7-3-2-5-14(16)19(26)21-15-6-4-8-20-18(15)24/h2-8H,9-13H2,1H3,(H,21,26) |
| PubChem CID | 4848 |
| ChEMBL | CHEMBL9967 |
| IUPHAR | 328 |
| BindingDB | 39341 |
| DrugBank | DB00670 |
Structure | 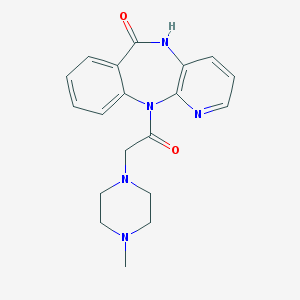 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| IC50 | 968.0 nM | PMID9986705 | BindingDB |
| IC50 | 968.2 nM | PMID9986705 | ChEMBL |
| Kb | 50000.0 nM | PMID2738887 | ChEMBL |
| Kd | 50118.7 nM | PMID2738887 | ChEMBL |
| Kd | 50119.0 nM | PMID2738887 | BindingDB |
| Ki | <10000.0 nM | PMID3272174 | PDSP,BindingDB |
| Ki | 17.3 nM | PMID2746503 | BindingDB |
| Ki | 173.78 nM | PMID18595721 | BindingDB,ChEMBL |
| Ki | 199.526 nM | PMID8016895 | PDSP |
| Ki | 199.526 - 1000.0 nM | PMID12049493, PMID2704370, PMID7925952, PMID9454790, PMID16188951, PMID1994002, PMID9113359, PMID2043926 | IUPHAR |
| Ki | 223.87 nM | PMID1994002 | BindingDB |
| Ki | 223.872 nM | PMID1994002 | PDSP |
| Ki | 251.0 nM | , Bioorg. Med. Chem. Lett., (1996) 6:7:785 | BindingDB,ChEMBL |
| Ki | 270.0 nM | PMID1346637, PMID7853341 | PDSP,BindingDB,ChEMBL |
| Ki | 374.0 nM | PMID9454790 | PDSP,BindingDB |
| Ki | 459.0 nM | PMID2746503 | BindingDB |
| Ki | 470.0 nM | PMID9121349 | PDSP |
| Ki | 524.0 nM | PMID23379472 | BindingDB,ChEMBL |
| Ki | 906.0 nM | PMID2704370 | PDSP,BindingDB |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417