

You can:
| Name | 5-hydroxytryptamine receptor 1B |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | HTR1B |
| Synonym | 5-HT1B receptor 5-HT-1B 5-HT-1D-beta 5-HT1B Serotonin 1D beta receptor [ Show all ] |
| Disease | Chronic schizophrenics Major depressive disorder Migraine headaches Mood disorder Psychotic disorders [ Show all ] |
| Length | 390 |
| Amino acid sequence | MEEPGAQCAPPPPAGSETWVPQANLSSAPSQNCSAKDYIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASILHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFFWRQAKAEEEVSECVVNTDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRILKQTPNRTGKRLTRAQLITDSPGSTSSVTSINSRVPDVPSESGSPVYVNQVKVRVSDALLEKKKLMAARERKATKTLGIILGAFIVCWLPFFIISLVMPICKDACWFHLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFKCTS |
| UniProt | P28222 |
| Protein Data Bank | 4iar, 6g79, 5v54 |
| GPCR-HGmod model | P28222 |
| 3D structure model | This structure is from PDB ID 4iar. |
| BioLiP | BL0239857, BL0403524,BL0403525, BL0417722 |
| Therapeutic Target Database | T07806 |
| ChEMBL | CHEMBL1898 |
| IUPHAR | 2 |
| DrugBank | BE0000797 |
| Name | serotonin |
|---|---|
| Molecular formula | C10H12N2O |
| IUPAC name | 3-(2-aminoethyl)-1H-indol-5-ol |
| Molecular weight | 176.219 |
| Hydrogen bond acceptor | 2 |
| Hydrogen bond donor | 3 |
| XlogP | 0.2 |
| Synonyms | NCGC00015525-08 Bio1_000450 PDSP1_001512 BPBio1_001079 CCG-204696 [ Show all ] |
| Inchi Key | QZAYGJVTTNCVMB-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C10H12N2O/c11-4-3-7-6-12-10-2-1-8(13)5-9(7)10/h1-2,5-6,12-13H,3-4,11H2 |
| PubChem CID | 5202 |
| ChEMBL | CHEMBL39 |
| IUPHAR | 5 |
| BindingDB | 10755 |
| DrugBank | DB08839 |
Structure | 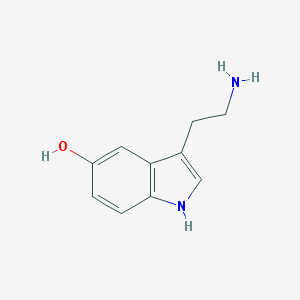 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| EC50 | 4.67 nM | PMID9871775 | ChEMBL |
| EC50 | 4.7 nM | PMID9871775 | BindingDB |
| EC50 | 4.82 nM | PMID8960551 | BindingDB,ChEMBL |
| EC50 | 30.0 nM | PMID2374139 | BindingDB,ChEMBL |
| IA | 1.0 - | PMID15887956 | ChEMBL |
| IC50 | 5.0 nM | PMID1447752 | BindingDB,ChEMBL |
| IC50 | 6.5 nM | PMID23466604 | ChEMBL |
| IC50 | 12.0 nM | PMID23582449 | ChEMBL |
| IC50 | 16.0 nM | PMID18588282 | ChEMBL |
| IC50 | 29.0 nM | PMID18983139 | ChEMBL |
| Intrinsic activity | 1.0 % | PMID9548813 | ChEMBL |
| Kd | 3.98107 - 31.6228 nM | PMID1565658, PMID8863519 | IUPHAR |
| Ki | 0.57 nM | PMID7984267 | PDSP,BindingDB |
| Ki | 0.59 nM | PMID1315531 | PDSP,BindingDB |
| Ki | 1.0 - 39.8107 nM | PMID10431754, PMID1565658, PMID9208142, PMID9776361, PMID11040052, PMID8957260 | IUPHAR |
| Ki | 1.09 nM | PMID7984267 | PDSP,BindingDB |
| Ki | 1.1 nM | PMID18507369 | PDSP |
| Ki | 1.1 nM | PMID18507369 | BindingDB,ChEMBL |
| Ki | 3.9 nM | PMID1565658 | PDSP,BindingDB |
| Ki | 3.98 nM | PMID7984267 | PDSP,BindingDB |
| Ki | 4.0 nM | PMID8071931, PMID23466604, PMID8568822 | BindingDB,ChEMBL |
| Ki | 4.07 nM | PMID9776361 | PDSP,BindingDB |
| Ki | 4.26 nM | PMID7984267 | PDSP,BindingDB |
| Ki | 4.8 nM | PMID12519065 | BindingDB,ChEMBL |
| Ki | 5.012 nM | PMID14613313 | ChEMBL |
| Ki | 6.76 nM | PMID8960551, PMID9871775 | ChEMBL |
| Ki | 6.8 nM | PMID8960551, PMID9871775 | BindingDB |
| Ki | 7.1 nM | PMID7658447 | BindingDB,ChEMBL |
| Ki | 7.94 nM | PMID9303567 | PDSP,BindingDB |
| Ki | 9.0 nM | PMID9303569 | PDSP,BindingDB |
| Ki | 14.0 nM | PMID21726069 | BindingDB,ChEMBL |
| Ki | 15.84 nM | PMID7984267 | PDSP,BindingDB |
| Ki | 18.0 nM | PMID18983139 | ChEMBL |
| Ki | 38.0 nM | PMID9303569 | PDSP,BindingDB |
| Simulation | 219.0 % | PMID9548813 | ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417