

You can:
| Name | 5-hydroxytryptamine receptor 1A |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | HTR1A |
| Synonym | 5-HT-1A 5-HT1A serotonin receptor 1A 5-HT1A receptor 5-hydroxytryptamine (serotonin) receptor 1A, G protein-coupled [ Show all ] |
| Disease | Urinary incontinence Generalized anxiety disorder Generalized anxiety disorder; Social phobia Hypertension Hypoactive sexual desire disorder [ Show all ] |
| Length | 422 |
| Amino acid sequence | MDVLSPGQGNNTTSPPAPFETGGNTTGISDVTVSYQVITSLLLGTLIFCAVLGNACVVAAIALERSLQNVANYLIGSLAVTDLMVSVLVLPMAALYQVLNKWTLGQVTCDLFIALDVLCCTSSILHLCAIALDRYWAITDPIDYVNKRTPRRAAALISLTWLIGFLISIPPMLGWRTPEDRSDPDACTISKDHGYTIYSTFGAFYIPLLLMLVLYGRIFRAARFRIRKTVKKVEKTGADTRHGASPAPQPKKSVNGESGSRNWRLGVESKAGGALCANGAVRQGDDGAALEVIEVHRVGNSKEHLPLPSEAGPTPCAPASFERKNERNAEAKRKMALARERKTVKTLGIIMGTFILCWLPFFIVALVLPFCESSCHMPTLLGAIINWLGYSNSLLNPVIYAYFNKDFQNAFKKIIKCKFCRQ |
| UniProt | P08908 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | P08908 |
| 3D structure model | This predicted structure model is from GPCR-EXP P08908. |
| BioLiP | N/A |
| Therapeutic Target Database | T78709 |
| ChEMBL | CHEMBL214 |
| IUPHAR | 1 |
| DrugBank | BE0000291 |
| Name | serotonin |
|---|---|
| Molecular formula | C10H12N2O |
| IUPAC name | 3-(2-aminoethyl)-1H-indol-5-ol |
| Molecular weight | 176.219 |
| Hydrogen bond acceptor | 2 |
| Hydrogen bond donor | 3 |
| XlogP | 0.2 |
| Synonyms | Bio1_000450 PDSP1_001512 BPBio1_001079 CCG-204696 Thrombocytin [ Show all ] |
| Inchi Key | QZAYGJVTTNCVMB-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C10H12N2O/c11-4-3-7-6-12-10-2-1-8(13)5-9(7)10/h1-2,5-6,12-13H,3-4,11H2 |
| PubChem CID | 5202 |
| ChEMBL | CHEMBL39 |
| IUPHAR | 5 |
| BindingDB | 10755 |
| DrugBank | DB08839 |
Structure | 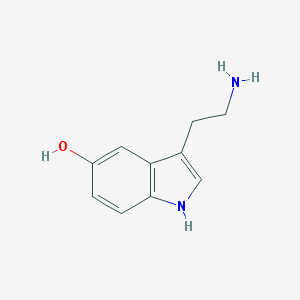 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| Activity | 99.6 % | PMID23675993 | ChEMBL |
| Activity | 99.7 % | PMID27487565 | ChEMBL |
| Activity | 100.0 % | PMID20363635 | ChEMBL |
| EC50 | 0.5 nM | PMID16392798 | BindingDB |
| EC50 | 0.5 nM | PMID16392798 | ChEMBL |
| EC50 | 0.7 nM | PMID16458504 | ChEMBL |
| EC50 | 0.7 nM | PMID16458504 | BindingDB |
| EC50 | 1.0 nM | PMID16392798 | BindingDB,ChEMBL |
| EC50 | 3.0 nM | PMID25308766 | ChEMBL |
| EC50 | 3.0 nM | PMID25308766 | BindingDB |
| EC50 | 3.7 nM | PMID12954071 | BindingDB,ChEMBL |
| EC50 | 3.75 nM | PMID23332346 | ChEMBL |
| EC50 | 3.8 nM | PMID23332346 | BindingDB |
| EC50 | 8.71 nM | PMID14613313 | ChEMBL |
| EC50 | 10.0 nM | PMID1447752 | BindingDB,ChEMBL |
| EC50 | 15.0 nM | PMID27487565 | BindingDB |
| EC50 | 15.33 nM | PMID27487565 | ChEMBL |
| EC50 | 27.54 nM | PMID26081758 | ChEMBL |
| EC50 | 28.0 nM | PMID26081758 | BindingDB |
| EC50 | 39.0 nM | PMID23675993 | ChEMBL |
| EC50 | 43.0 nM | , Bioorg. Med. Chem. Lett., (1995) 5:20:2391 | BindingDB,ChEMBL |
| EC50 | 147.0 nM | PMID20041669 | BindingDB,ChEMBL |
| EC50 | 150.0 nM | PMID19559623 | BindingDB,ChEMBL |
| EC50 | 169.82 nM | PMID26081758 | ChEMBL |
| EC50 | 170.0 nM | PMID26081758 | BindingDB |
| EC50 | 229.09 nM | PMID17803293 | ChEMBL |
| Emax | 100.0 % | PMID20041669, PMID23332346 | ChEMBL |
| Emax | 224.0 - | PMID14613313 | ChEMBL |
| FC | 6.4 - | PMID26081758 | ChEMBL |
| IA | 1.0 - | PMID15887956 | ChEMBL |
| IC50 | 1.2 nM | PMID12954071 | BindingDB,ChEMBL |
| IC50 | 2.0 nM | PMID2918500 | BindingDB,ChEMBL |
| IC50 | 5.2 nM | PMID1447752 | BindingDB,ChEMBL |
| IC50 | 5.6 nM | PMID2918500 | BindingDB,ChEMBL |
| IC50 | 6.0 nM | PMID2918500 | BindingDB,ChEMBL |
| Inhibition | 100.0 % | PMID23332346 | ChEMBL |
| Intrinsic activity | 1.0 - | PMID18433113, PMID14613313 | ChEMBL |
| Ki | 0.199526 - 0.794328 nM | PMID1386736, PMID9760039, PMID10431754, PMID9550290, PMID9205951, PMID15628665 | IUPHAR |
| Ki | 0.26 nM | PMID7984267 | PDSP,BindingDB |
| Ki | 0.62 nM | PMID22748706 | BindingDB,ChEMBL |
| Ki | 0.72 nM | PMID7855217 | PDSP |
| Ki | 0.79 nM | PMID7984267 | PDSP,BindingDB |
| Ki | 0.9 nM | PMID23332346 | ChEMBL |
| Ki | 0.9 nM | PMID23332346 | BindingDB |
| Ki | 1.2 nM | PMID27543433, PMID12954071 | PDSP,BindingDB |
| Ki | 1.25 nM | PMID7984267 | PDSP,BindingDB |
| Ki | 1.259 nM | PMID18433113 | ChEMBL |
| Ki | 1.3 nM | PMID18433113, PMID9686407 | PDSP,BindingDB |
| Ki | 1.68 nM | Hoyer et al., PMID1986 | PDSP |
| Ki | 1.7 nM | PMID8071931 | BindingDB,ChEMBL |
| Ki | 1.75 nM | Hoyer et al., PMID1986 | PDSP |
| Ki | 2.08 nM | PMID25128671 | ChEMBL |
| Ki | 2.1 nM | PMID25128671 | BindingDB |
| Ki | 2.48 nM | PMID8960551, PMID9871775 | BindingDB,ChEMBL |
| Ki | 2.5 nM | PMID7658447, PMID9871775 | BindingDB,ChEMBL |
| Ki | 2.8 nM | PMID2918500 | BindingDB,ChEMBL |
| Ki | 3.0 nM | PMID25048712, PMID2918500 | BindingDB,ChEMBL |
| Ki | 3.98 nM | PMID7984267, PMID8461029 | PDSP,BindingDB |
| Ki | 3.981 nM | PMID14613313 | ChEMBL |
| Ki | 9.5 nM | PMID24161678 | ChEMBL |
| Ki | 10.0 nM | PMID22738316 | BindingDB |
| Ki | 10.0 nM | PMID23488743 | BindingDB |
| Ki | 12.58 nM | PMID7984267 | PDSP,BindingDB |
| Ki | >50.0 nM | PMID20363635 | ChEMBL |
| Ki | 102.3 nM | PMID8461029 | PDSP,BindingDB |
| Ki | 102.32 nM | PMID7984267 | PDSP,BindingDB |
| Ki | 166.0 nM | PMID8155646 | PDSP |
| Ki | 10000.0 nM | PMID18433113 | BindingDB,ChEMBL |
| Max | 100.0 % | PMID10514291, PMID10425105 | ChEMBL |
| pD2 | 7.3 - | PMID18817363, PMID10425105, PMID23252794, PMID10514291 | ChEMBL |
| pKD | 8.5 - | PMID8515429 | ChEMBL |
| Stimulation | 126.0 % | PMID14613313 | ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417