

You can:
| Name | Histamine H3 receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | HRH3 |
| Synonym | HH3R H3R H3 receptor GPCR97 G-protein coupled receptor 97 |
| Disease | Obese insulin-resistant disorders Excessive daytime sleepiness Sleep disorders Schizophrenia Pain [ Show all ] |
| Length | 445 |
| Amino acid sequence | MERAPPDGPLNASGALAGEAAAAGGARGFSAAWTAVLAALMALLIVATVLGNALVMLAFVADSSLRTQNNFFLLNLAISDFLVGAFCIPLYVPYVLTGRWTFGRGLCKLWLVVDYLLCTSSAFNIVLISYDRFLSVTRAVSYRAQQGDTRRAVRKMLLVWVLAFLLYGPAILSWEYLSGGSSIPEGHCYAEFFYNWYFLITASTLEFFTPFLSVTFFNLSIYLNIQRRTRLRLDGAREAAGPEPPPEAQPSPPPPPGCWGCWQKGHGEAMPLHRYGVGEAAVGAEAGEATLGGGGGGGSVASPTSSSGSSSRGTERPRSLKRGSKPSASSASLEKRMKMVSQSFTQRFRLSRDRKVAKSLAVIVSIFGLCWAPYTLLMIIRAACHGHCVPDYWYETSFWLLWANSAVNPVLYPLCHHSFRRAFTKLLCPQKLKIQPHSSLEHCWK |
| UniProt | Q9Y5N1 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | Q9Y5N1 |
| 3D structure model | This predicted structure model is from GPCR-EXP Q9Y5N1. |
| BioLiP | N/A |
| Therapeutic Target Database | T64765 |
| ChEMBL | CHEMBL264 |
| IUPHAR | 264 |
| DrugBank | BE0000968 |
| Name | imetit |
|---|---|
| Molecular formula | C6H10N4S |
| IUPAC name | 2-(1H-imidazol-5-yl)ethyl carbamimidothioate |
| Molecular weight | 170.234 |
| Hydrogen bond acceptor | 3 |
| Hydrogen bond donor | 3 |
| XlogP | 0.2 |
| Synonyms | C6H10N4S D02RAN NCGC00015536-01 S-(2-(4-Imidazolyl)ethyl)isothiourea 677MJ4VPZC [ Show all ] |
| Inchi Key | PEHSVUKQDJULKE-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C6H10N4S/c7-6(8)11-2-1-5-3-9-4-10-5/h3-4H,1-2H2,(H3,7,8)(H,9,10) |
| PubChem CID | 3692 |
| ChEMBL | CHEMBL19439 |
| IUPHAR | 1250 |
| BindingDB | 22911 |
| DrugBank | N/A |
Structure | 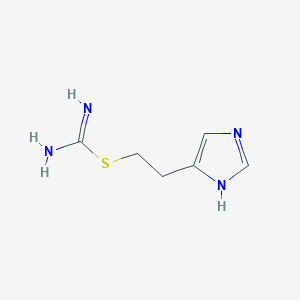 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| Activity | 91.0 % | PMID20384344 | ChEMBL |
| EC50 | 0.1259 nM | PMID21044842 | ChEMBL |
| EC50 | 1.9 nM | PMID12723960 | BindingDB,ChEMBL |
| EC50 | 2.4 nM | PMID20384344 | BindingDB,ChEMBL |
| Intrinsic activity | 1.0 - | PMID21044842 | ChEMBL |
| Ki | <10000.0 nM | PMID10347254 | PDSP,BindingDB |
| Ki | 0.199526 - 1.58489 nM | PMID10869375, PMID11284713, PMID11714875, PMID12393057, PMID11179434 | IUPHAR |
| Ki | 0.3 nM | PMID11179434 | PDSP,BindingDB |
| Ki | 0.57 nM | PMID20384344 | BindingDB |
| Ki | 0.57 nM | PMID20384344 | PDSP |
| Ki | 0.57 nM | PMID20384344 | ChEMBL |
| Ki | 0.6 nM | PMID10869375 | PDSP,BindingDB |
| Ki | 0.7 nM | PMID21498080 | BindingDB,ChEMBL |
| Ki | 1.58 nM | PMID19414267, PMID22003888 | BindingDB |
| Ki | 1.585 nM | PMID19414267, PMID21044842, PMID22003888 | ChEMBL |
| Ki | 1.62 nM | PMID11714875 | BindingDB |
| Ki | 1.62181 nM | PMID11714875 | PDSP |
| Ki | 2.0 nM | PMID15947036 | BindingDB |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417