

You can:
| Name | Metabotropic glutamate receptor 5 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | GRM5 |
| Synonym | mGlu5 receptor glutamate receptor GPRC1E mGluR5 |
| Disease | Central nervous system disease Chronic neuropathic pain Fragile X syndrome Major depressive disorder; GERD; Chronic neuropathic pain Autism [ Show all ] |
| Length | 1212 |
| Amino acid sequence | MVLLLILSVLLLKEDVRGSAQSSERRVVAHMPGDIIIGALFSVHHQPTVDKVHERKCGAVREQYGIQRVEAMLHTLERINSDPTLLPNITLGCEIRDSCWHSAVALEQSIEFIRDSLISSEEEEGLVRCVDGSSSSFRSKKPIVGVIGPGSSSVAIQVQNLLQLFNIPQIAYSATSMDLSDKTLFKYFMRVVPSDAQQARAMVDIVKRYNWTYVSAVHTEGNYGESGMEAFKDMSAKEGICIAHSYKIYSNAGEQSFDKLLKKLTSHLPKARVVACFCEGMTVRGLLMAMRRLGLAGEFLLLGSDGWADRYDVTDGYQREAVGGITIKLQSPDVKWFDDYYLKLRPETNHRNPWFQEFWQHRFQCRLEGFPQENSKYNKTCNSSLTLKTHHVQDSKMGFVINAIYSMAYGLHNMQMSLCPGYAGLCDAMKPIDGRKLLESLMKTNFTGVSGDTILFDENGDSPGRYEIMNFKEMGKDYFDYINVGSWDNGELKMDDDEVWSKKSNIIRSVCSEPCEKGQIKVIRKGEVSCCWTCTPCKENEYVFDEYTCKACQLGSWPTDDLTGCDLIPVQYLRWGDPEPIAAVVFACLGLLATLFVTVVFIIYRDTPVVKSSSRELCYIILAGICLGYLCTFCLIAKPKQIYCYLQRIGIGLSPAMSYSALVTKTNRIARILAGSKKKICTKKPRFMSACAQLVIAFILICIQLGIIVALFIMEPPDIMHDYPSIREVYLICNTTNLGVVTPLGYNGLLILSCTFYAFKTRNVPANFNEAKYIAFTMYTTCIIWLAFVPIYFGSNYKIITMCFSVSLSATVALGCMFVPKVYIILAKPERNVRSAFTTSTVVRMHVGDGKSSSAASRSSSLVNLWKRRGSSGETLRYKDRRLAQHKSEIECFTPKGSMGNGGRATMSSSNGKSVTWAQNEKSSRGQHLWQRLSIHINKKENPNQTAVIKPFPKSTESRGLGAGAGAGGSAGGVGATGGAGCAGAGPGGPESPDAGPKALYDVAEAEEHFPAPARPRSPSPISTLSHRAGSASRTDDDVPSLHSEPVARSSSSQGSLMEQISSVVTRFTANISELNSMMLSTAAPSPGVGAPLCSSYLIPKEIQLPTTMTTFAEIQPLPAIEVTGGAQPAAGAQAAGDAARESPAAGPEAAAAKPDLEELVALTPPSPFRDSVDSGSTTPNSPVSESALCIPSSPKYDTLIIRDYTQSSSSL |
| UniProt | P41594 |
| Protein Data Bank | 4oo9, 5cgc, 5cgd, 6ffh, 6ffi, 6n4x, 6n4y, 6n50, 6n51, 3lmk |
| GPCR-HGmod model | N/A |
| 3D structure model | This structure is from PDB ID 4oo9. |
| BioLiP | BL0176927,BL0176931, BL0176928,BL0176929,BL0176930, BL0281199, BL0322076, BL0322077, BL0407724, BL0438693, BL0438694, BL0438695,BL0438696,BL0438697, BL0407725, BL0438698,BL0438699 |
| Therapeutic Target Database | T99347 |
| ChEMBL | CHEMBL3227 |
| IUPHAR | 293 |
| DrugBank | BE0001192 |
| Name | MPEP |
|---|---|
| Molecular formula | C14H11N |
| IUPAC name | 2-methyl-6-(2-phenylethynyl)pyridine |
| Molecular weight | 193.249 |
| Hydrogen bond acceptor | 1 |
| Hydrogen bond donor | 0 |
| XlogP | 3.3 |
| Synonyms | 7VC0YVI27Y BRD-K60690191-003-02-1 DB-014655 Lopac-M-5435 NCGC00015682-03 [ Show all ] |
| Inchi Key | NEWKHUASLBMWRE-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C14H11N/c1-12-6-5-9-14(15-12)11-10-13-7-3-2-4-8-13/h2-9H,1H3 |
| PubChem CID | 3025961 |
| ChEMBL | CHEMBL66654 |
| IUPHAR | 1426 |
| BindingDB | 50084137 |
| DrugBank | N/A |
Structure | 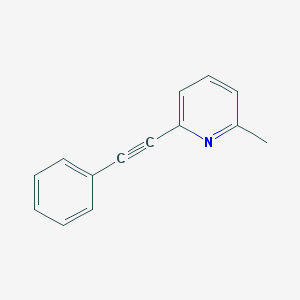 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| Bmax | 4.4 pmol | PMID23046966 | ChEMBL |
| IC50 | 1.5 nM | PMID17569516 | BindingDB,ChEMBL |
| IC50 | 2.0 nM | PMID12519057, PMID12565928 | BindingDB,ChEMBL |
| IC50 | 2.3 nM | PMID23357634 | BindingDB,ChEMBL |
| IC50 | 4.0 nM | PMID17196387 | BindingDB,ChEMBL |
| IC50 | 10.0 nM | PMID18304814 | BindingDB,ChEMBL |
| IC50 | 11.0 nM | PMID26014480, PMID12565928 | BindingDB,ChEMBL |
| IC50 | 11.2 nM | PMID26014480 | ChEMBL |
| IC50 | 19.9526 - 39.8107 nM | PMID11814808, PMID10530811 | IUPHAR |
| IC50 | 20.0 nM | PMID11814808 | BindingDB,ChEMBL |
| IC50 | 22.0 nM | PMID16839764, PMID16481165 | BindingDB,ChEMBL |
| IC50 | 29.0 nM | PMID17196387 | BindingDB,ChEMBL |
| IC50 | 32.0 nM | PMID26112438 | BindingDB,ChEMBL |
| IC50 | 36.0 nM | PMID10639281, PMID11814808 | BindingDB,ChEMBL |
| IC50 | 39.0 nM | PMID16678408, PMID16451073 | BindingDB,ChEMBL |
| Inhibition | <70.0 % | PMID19445453 | ChEMBL |
| Kd | 8.6 nM | PMID23046966 | BindingDB,ChEMBL |
| Ki | 3.3 nM | PMID16040814 | PDSP,BindingDB |
| Ki | 3.4 nM | PMID23357634 | BindingDB,ChEMBL |
| Ki | 3.5 nM | PMID26706173 | BindingDB,ChEMBL |
| Ki | 6.7 nM | PMID16040814 | PDSP,BindingDB |
| Ki | 10.0 nM | PMID26014480 | BindingDB |
| Ki | 10.4 nM | PMID26014480 | ChEMBL |
| Ki | 13.0 nM | PMID19445453 | BindingDB,ChEMBL |
| Ki | 20.0 nM | PMID17110115, PMID11814808 | PDSP,BindingDB,ChEMBL |
| Ki | 28.0 nM | PMID16040814 | PDSP,BindingDB |
| Ki | 36.0 nM | PMID10893301 | BindingDB,ChEMBL |
| Ki | 36.31 nM | PMID20483612 | BindingDB,ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417