

You can:
| Name | Alpha-1A adrenergic receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | ADRA1A |
| Synonym | alpha1a ADRA1C ADRA1L1 adrenergic alpha 1c receptor adrenergic receptor alpha 1c [ Show all ] |
| Disease | Urinary incontinence Benign prostatic hyperplasia Cognitive disorders Female sexual dysfunction Glaucoma [ Show all ] |
| Length | 466 |
| Amino acid sequence | MVFLSGNASDSSNCTQPPAPVNISKAILLGVILGGLILFGVLGNILVILSVACHRHLHSVTHYYIVNLAVADLLLTSTVLPFSAIFEVLGYWAFGRVFCNIWAAVDVLCCTASIMGLCIISIDRYIGVSYPLRYPTIVTQRRGLMALLCVWALSLVISIGPLFGWRQPAPEDETICQINEEPGYVLFSALGSFYLPLAIILVMYCRVYVVAKRESRGLKSGLKTDKSDSEQVTLRIHRKNAPAGGSGMASAKTKTHFSVRLLKFSREKKAAKTLGIVVGCFVLCWLPFFLVMPIGSFFPDFKPSETVFKIVFWLGYLNSCINPIIYPCSSQEFKKAFQNVLRIQCLCRKQSSKHALGYTLHPPSQAVEGQHKDMVRIPVGSRETFYRISKTDGVCEWKFFSSMPRGSARITVSKDQSSCTTARVRSKSFLQVCCCVGPSTPSLDKNHQVPTIKVHTISLSENGEEV |
| UniProt | P35348 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | P35348 |
| 3D structure model | This predicted structure model is from GPCR-EXP P35348. |
| BioLiP | N/A |
| Therapeutic Target Database | T92609 |
| ChEMBL | CHEMBL229 |
| IUPHAR | 22 |
| DrugBank | BE0000501 |
| Name | AC1NKXXY |
|---|---|
| Molecular formula | C17H19N3O |
| IUPAC name | 3-[N-(4,5-dihydro-1H-imidazol-3-ium-2-ylmethyl)-4-methylanilino]phenolate |
| Molecular weight | 281.359 |
| Hydrogen bond acceptor | 2 |
| Hydrogen bond donor | 2 |
| XlogP | 3.3 |
| Synonyms | MolPort-000-771-097 3-[N-(4,5-dihydro-1H-imidazol-3-ium-2-ylmethyl)-4-methylanilino]phenolate STL054299 AKOS005707227 MCULE-3004652227 [ Show all ] |
| Inchi Key | MRBDMNSDAVCSSF-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C17H19N3O/c1-13-5-7-14(8-6-13)20(12-17-18-9-10-19-17)15-3-2-4-16(21)11-15/h2-8,11,21H,9-10,12H2,1H3,(H,18,19) |
| PubChem CID | 4920903 |
| ChEMBL | CHEMBL597 |
| IUPHAR | 502 |
| BindingDB | 31046 |
| DrugBank | DB00692 |
Structure | 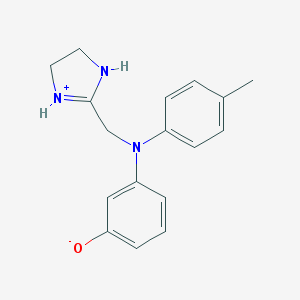 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| N/A | N/A | DrugBank | |
| IC50 | 1.1 nM | PMID26005522 | BindingDB,ChEMBL |
| Kd | 32.0 nM | PMID6142954 | BindingDB |
| Kd | 32.36 nM | PMID6142954 | ChEMBL |
| Ki | 0.6 nM | PMID26005522 | ChEMBL |
| Ki | 0.6 nM | PMID26005522 | BindingDB |
| Ki | 1.4 nM | PMID8396931 | BindingDB |
| Ki | 2.51188 nM | PMID7651358 | IUPHAR |
| Ki | 3.4 nM | PMID9400006 | BindingDB |
| Ki | 4.1 nM | PMID16153828 | BindingDB |
| Ki | 4.12 nM | PMID16153828 | ChEMBL |
| Ki | 14.92 nM | PMID2904784 | BindingDB |
| Ki | 15.0 nM | PMID6149136 | BindingDB |
| Ki | 19.95 nM | PMID7815325 | BindingDB |
| Ki | 31.62 nM | PMID7815325 | BindingDB |
| Ki | 199.52 nM | PMID7815325 | BindingDB |
| Selectivity | 4.8 - | PMID6133953 | ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417