

You can:
| Name | Adenosine receptor A1 |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Adora1 |
| Synonym | A1 receptor A1-AR A1R adenosine receptor A1 RDC7 |
| Disease | N/A for non-human GPCRs |
| Length | 326 |
| Amino acid sequence | MPPYISAFQAAYIGIEVLIALVSVPGNVLVIWAVKVNQALRDATFCFIVSLAVADVAVGALVIPLAILINIGPQTYFHTCLMVACPVLILTQSSILALLAIAVDRYLRVKIPLRYKTVVTQRRAAVAIAGCWILSLVVGLTPMFGWNNLSVVEQDWRANGSVGEPVIKCEFEKVISMEYMVYFNFFVWVLPPLLLMVLIYLEVFYLIRKQLNKKVSASSGDPQKYYGKELKIAKSLALILFLFALSWLPLHILNCITLFCPTCQKPSILIYIAIFLTHGNSAMNPIVYAFRIHKFRVTFLKIWNDHFRCQPKPPIDEDLPEEKAED |
| UniProt | P25099 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL318 |
| IUPHAR | 18 |
| DrugBank | N/A |
| Name | 1,3-Dipropylxanthine |
|---|---|
| Molecular formula | C11H16N4O2 |
| IUPAC name | 1,3-dipropyl-7H-purine-2,6-dione |
| Molecular weight | 236.275 |
| Hydrogen bond acceptor | 3 |
| Hydrogen bond donor | 1 |
| XlogP | 1.9 |
| Synonyms | ZINC4654890 1,3-dipropyl-7H-purine-2,6-dione AC1L53IX CAS_169317 DTXSID00185447 [ Show all ] |
| Inchi Key | MJVIGUCNSRXAFO-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C11H16N4O2/c1-3-5-14-9-8(12-7-13-9)10(16)15(6-4-2)11(14)17/h7H,3-6H2,1-2H3,(H,12,13) |
| PubChem CID | 169317 |
| ChEMBL | CHEMBL157655 |
| IUPHAR | 427 |
| BindingDB | 82032 |
| DrugBank | N/A |
Structure | 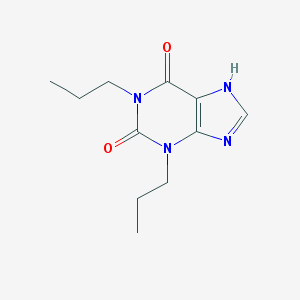 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| IC50 | 1500.0 nM | PMID6090665 | BindingDB,ChEMBL |
| Kb | 710.0 nM | PMID3346878 | ChEMBL |
| Ki | 450.0 nM | PMID3010074, PMID2754711, PMID1613758, PMID1548682, PMID2795597 | BindingDB,ChEMBL |
| Ki | 700.0 nM | PMID8410976, PMID3172141, PMID3806581, PMID8230124, PMID8182711, PMID2984420 | BindingDB,ChEMBL |
| Ki | 710.0 nM | PMID2724296 | BindingDB,ChEMBL |
| Ki | 1600.0 nM | PMID2724296 | BindingDB,ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417