

You can:
| Name | Delta-type opioid receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | OPRD1 |
| Synonym | D-OR-1 DOR opioid receptor OP1 DOP [ Show all ] |
| Disease | Cough Overactive bladder disorder Bladder disease Moderate-to-severe pain Diarrhea-predominant IBS [ Show all ] |
| Length | 372 |
| Amino acid sequence | MEPAPSAGAELQPPLFANASDAYPSACPSAGANASGPPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCGRPDPSSFSRAREATARERVTACTPSDGPGGGAAA |
| UniProt | P41143 |
| Protein Data Bank | 4rwd, 4rwa, 4n6h |
| GPCR-HGmod model | P41143 |
| 3D structure model | This structure is from PDB ID 4rwd. |
| BioLiP | BL0303696,BL0303697, BL0265712, BL0265705,BL0265706,BL0265707,, BL0303699,BL0303701, BL0303698,BL0303700 |
| Therapeutic Target Database | T58992 |
| ChEMBL | CHEMBL236 |
| IUPHAR | 317 |
| DrugBank | BE0000420 |
| Name | CHEMBL294616 |
|---|---|
| Molecular formula | C30H39N5O7S2 |
| IUPAC name | (4S,7R,13S)-13-[[(2S)-2-amino-3-(4-hydroxyphenyl)propanoyl]amino]-7-benzyl-3,3,14,14-tetramethyl-6,9,12-trioxo-1,2-dithia-5,8,11-triazacyclotetradecane-4-carboxylic acid |
| Molecular weight | 645.79 |
| Hydrogen bond acceptor | 10 |
| Hydrogen bond donor | 7 |
| XlogP | -0.6 |
| Synonyms | BDBM50001683 MCMMCRYPQBNCPH-DVKRWUGUSA-N 13-[2-Amino-3-(4-hydroxy-phenyl)-propionylamino]-7-benzyl-3,3,14,14-tetramethyl-6,9,12-trioxo-1,2-dithia-5,8,11-triaza-cyclotetradecane-4-carboxylic acid |
| Inchi Key | MCMMCRYPQBNCPH-DVKRWUGUSA-N |
| Inchi ID | InChI=1S/C30H39N5O7S2/c1-29(2)23(34-25(38)20(31)14-18-10-12-19(36)13-11-18)27(40)32-16-22(37)33-21(15-17-8-6-5-7-9-17)26(39)35-24(28(41)42)30(3,4)44-43-29/h5-13,20-21,23-24,36H,14-16,31H2,1-4H3,(H,32,40)(H,33,37)(H,34,38)(H,35,39)(H,41,42)/t20-,21+,23-,24-/m0/s1 |
| PubChem CID | 44299404 |
| ChEMBL | CHEMBL294616 |
| IUPHAR | N/A |
| BindingDB | 50001683 |
| DrugBank | N/A |
Structure | 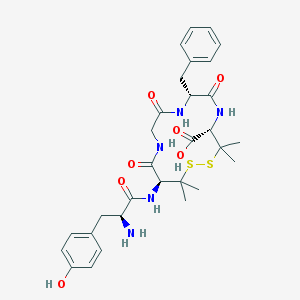 |
| Lipinski's druglikeness | This ligand has more than 5 hydrogen bond donor. This ligand is heavier than 500 daltons. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| EC50 | 0.14 nM | PMID21978284 | ChEMBL |
| EC50 | 0.14 nM | PMID21978284 | BindingDB |
| EC50 | 0.557 nM | PMID23477419 | BindingDB |
| EC50 | 0.557 nM | PMID23477419 | ChEMBL |
| EC50 | 0.8 nM | PMID25559207 | BindingDB |
| EC50 | 0.8 nM | PMID25559207 | ChEMBL |
| EC50 | 1.3 nM | PMID12672258 | BindingDB,ChEMBL |
| EC50 | 1.585 nM | PMID18821747, PMID16686530, PMID17516639, PMID20560643, PMID21128594, PMID16509592 | ChEMBL |
| EC50 | 1.6 nM | PMID18821747, PMID16686530, PMID17516639, PMID20560643, PMID21128594, PMID16509592, PMID18266313 | BindingDB,ChEMBL |
| EC50 | 2.951 nM | PMID19473027 | ChEMBL |
| EC50 | 3.0 nM | PMID19473027 | BindingDB |
| EC50 | 6.34 nM | PMID23659286 | ChEMBL |
| EC50 | 9.29 nM | PMID25248680 | ChEMBL |
| EC50 | 9.3 nM | PMID25248680 | BindingDB |
| EC50 | 15.0 nM | PMID19683449 | BindingDB,ChEMBL |
| EC50 | 15.85 nM | PMID26005537 | ChEMBL |
| EC50 | 16.0 nM | PMID26005537, PMID18637671 | BindingDB,ChEMBL |
| ED50 | 6.81 nM | PMID22325949 | ChEMBL |
| Emax | 69.0 % | PMID18821747, PMID16686530, PMID17516639, PMID21978284, PMID20560643, PMID21128594, PMID16509592, PMID18266313 | ChEMBL |
| Emax | 76.0 % | PMID19683449 | ChEMBL |
| Emax | 87.4 % | PMID23477419 | ChEMBL |
| Emax | 90.0 % | PMID1320122 | ChEMBL |
| Emax | 169.0 % | PMID19473027 | ChEMBL |
| Emax | 2200.0 % | PMID23659286 | ChEMBL |
| IC50 | 0.14 nM | PMID20599386 | BindingDB |
| IC50 | 0.14 nM | PMID20599386 | ChEMBL |
| IC50 | 0.9 nM | PMID25087049 | ChEMBL |
| IC50 | 0.9 nM | PMID25087049 | BindingDB |
| IC50 | 1.1 nM | PMID16989522, PMID17451272 | BindingDB |
| IC50 | 1.12 nM | PMID16989522, PMID17451272 | ChEMBL |
| IC50 | 1.2 nM | PMID18039010, PMID20426456, PMID17388627 | BindingDB,ChEMBL |
| IC50 | 1.8 nM | PMID7853350, PMID8071934 | BindingDB,ChEMBL |
| IC50 | 3.5 nM | PMID11906279 | ChEMBL |
| IC50 | 4.1 nM | PMID8831778 | BindingDB,ChEMBL |
| IC50 | 10.0 nM | PMID18588282 | ChEMBL |
| IC50 | 5000.0 nM | PMID7996538 | BindingDB,ChEMBL |
| IC50 | 7300.0 nM | PMID1320122 | BindingDB,ChEMBL |
| Imax | 69.0 % | PMID20599386 | ChEMBL |
| Ki | 0.5 nM | PMID20617791 | ChEMBL |
| Ki | 0.5 nM | PMID20617791 | BindingDB |
| Ki | 1.2 nM | PMID21866885 | BindingDB |
| Ki | 1.24 nM | PMID21866885 | ChEMBL |
| Ki | 1.39 nM | PMID21621410 | ChEMBL |
| Ki | 1.4 nM | PMID25513968, PMID21621410 | BindingDB |
| Ki | 1.41 nM | PMID25513968 | ChEMBL |
| Ki | 1.6 nM | PMID20560643 | BindingDB,ChEMBL |
| Ki | 1.7 nM | PMID12672258 | BindingDB,ChEMBL |
| Ki | 1.77 nM | PMID21621410 | ChEMBL |
| Ki | 1.8 nM | PMID21621410 | BindingDB |
| Ki | 2.2 nM | PMID12166947, PMID9651168 | BindingDB,ChEMBL |
| Ki | 2.9 nM | PMID1315868 | BindingDB,ChEMBL |
| Ki | 3.3 nM | PMID22995061, PMID25051243 | BindingDB,ChEMBL |
| Ki | 3.98 nM | PMID1333014, PMID1315870 | ChEMBL |
| Ki | 4.0 nM | PMID1333014, PMID1315870 | BindingDB |
| logEC50 | -8.8 - | PMID18266313 | ChEMBL |
| Selectivity ratio | 3270.0 - | PMID1320122 | ChEMBL |
| Stimulation | 100.0 % | PMID12672258 | ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417