Methods
Starting from an RNA sequence, DRfold2 first employs a pretrained RCLM to embed
the query into sequence and pair representations. Here, RCLM achieves superior sequence
pattern recognition through training on large-scale unsupervised sequence data using a
composite likelihood 26 maximization approach. These sequence and pair representations are
processed by a set of RNA Transformer Blocks, which produce necessary representations for structure folding.
DRfold2 then generates RNA conformations through a Denoising RNA Structure Module (DRSM) in an end-to-end fashion.
The final RNA models are obtained through a post-processing CSOR protocol that is designed to select and refine conformation decoys generated from a pool of checkpoints.
(see Fig. 1).
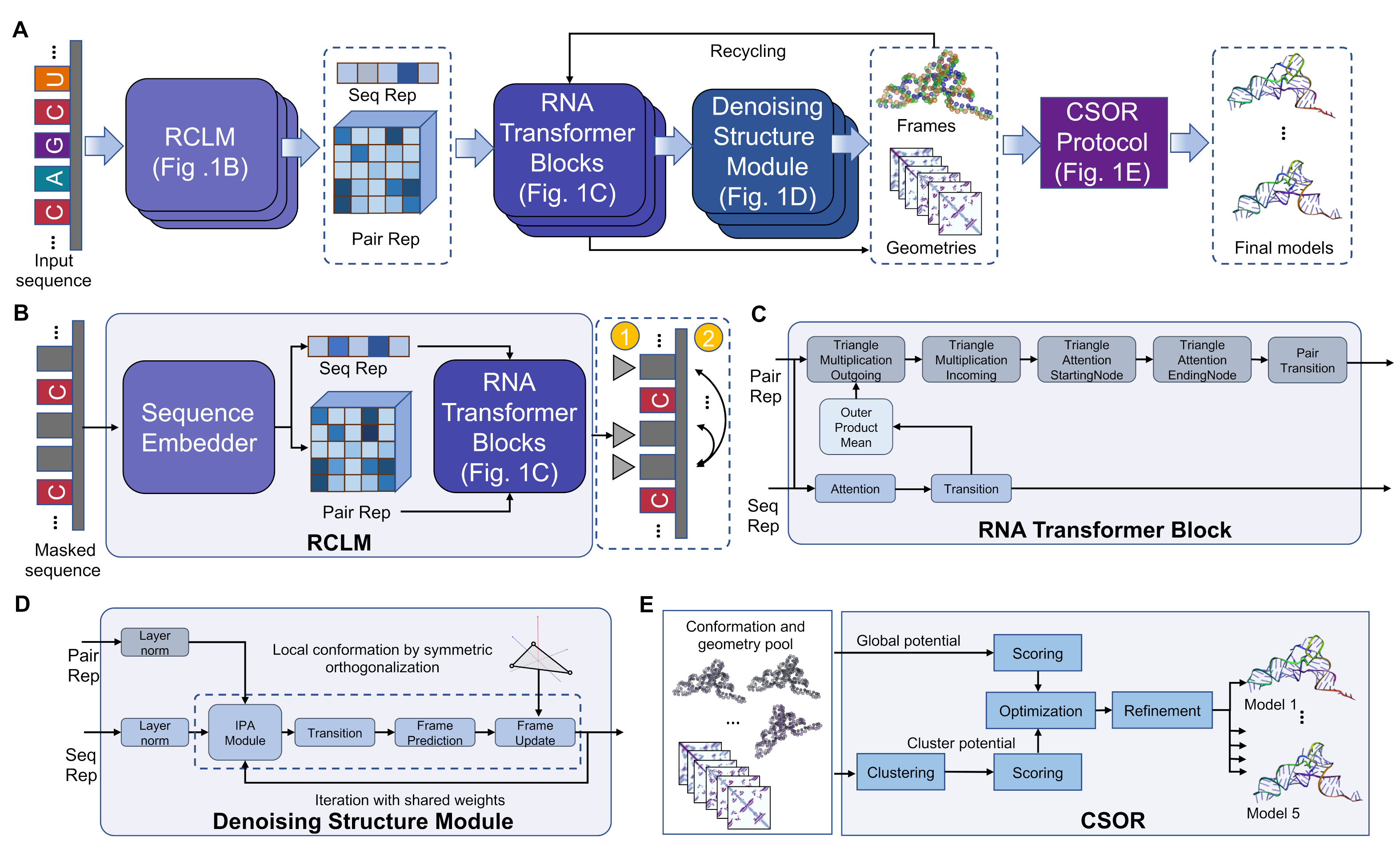 Figure 1. Pipeline of DRfold2.
Figure 1. Pipeline of DRfold2.
Performance
DRfold2 was tested on a set of 28 targets collected from PDB. The performance of DRfold2 is significantly better
than existing RNA structure predictors. Fig. 2 compares the performance evaluated by RMSD and TM-score of P atoms.
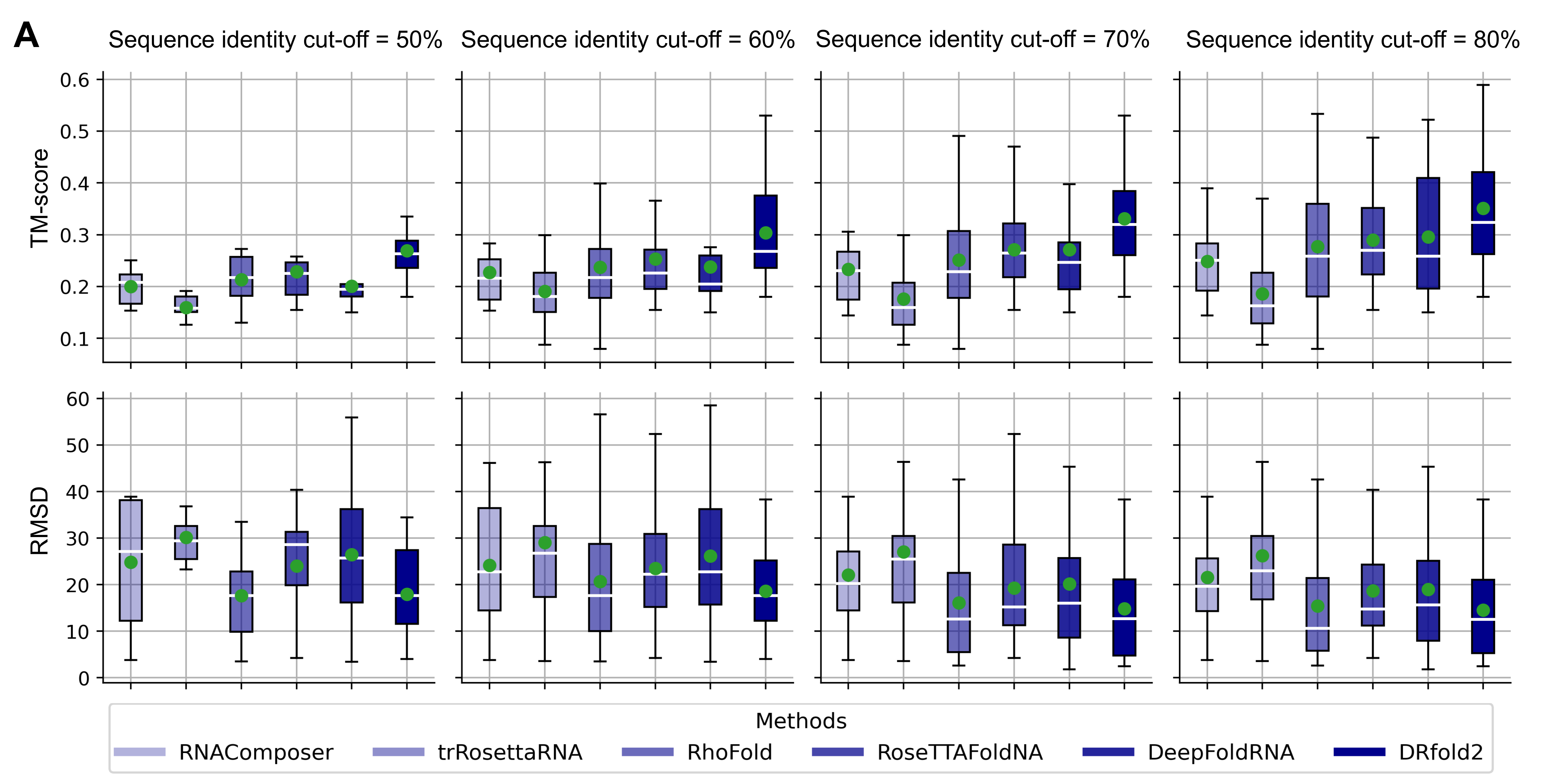 Figure 2. Performance comparisons between DRfold2 and
other RNA structure prediction methods.
Figure 2. Performance comparisons between DRfold2 and
other RNA structure prediction methods.
Server inputs
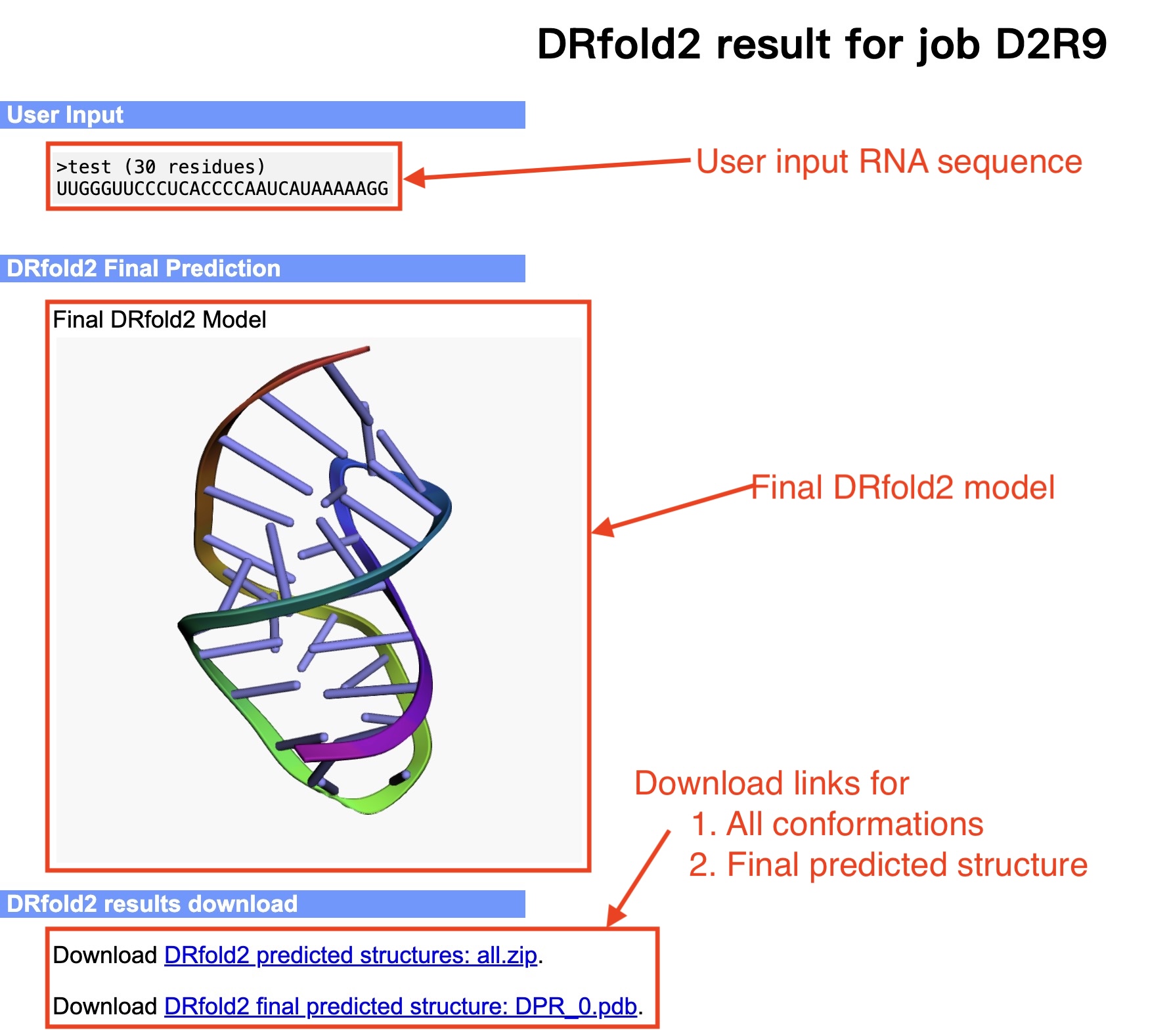
The user needs to paste the RNA sequence into the input box,
or upload the sequence of the query RNA using the "Choose file" button (see Fig. 3).
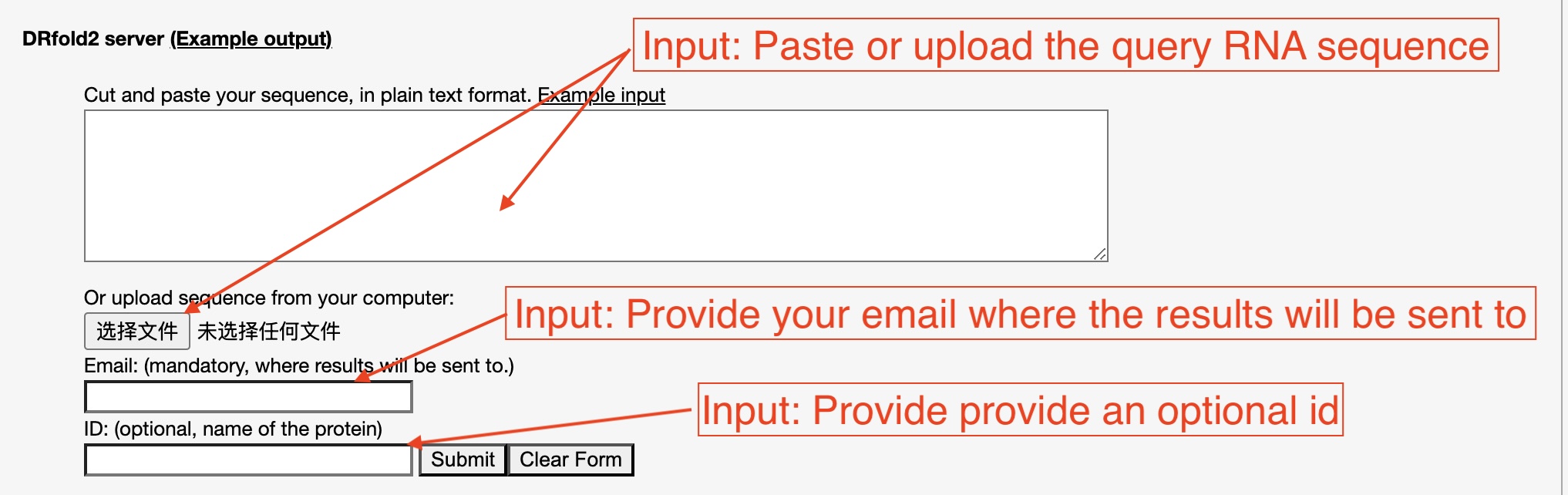
Figure 3. Illustration of input for the DRfold2 server.
Server outputs
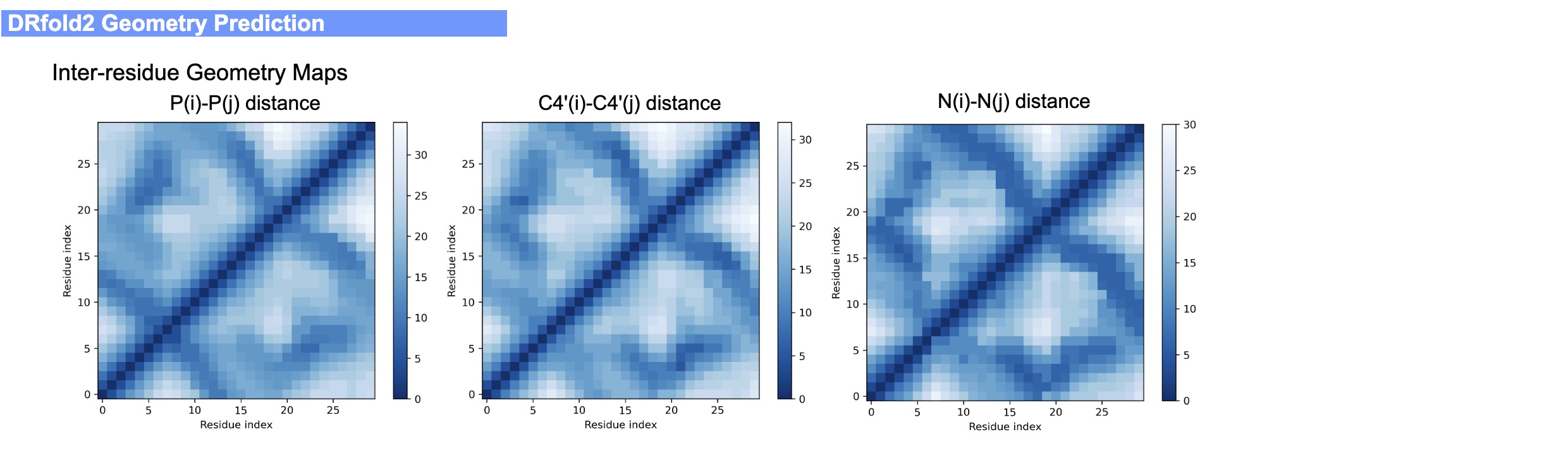
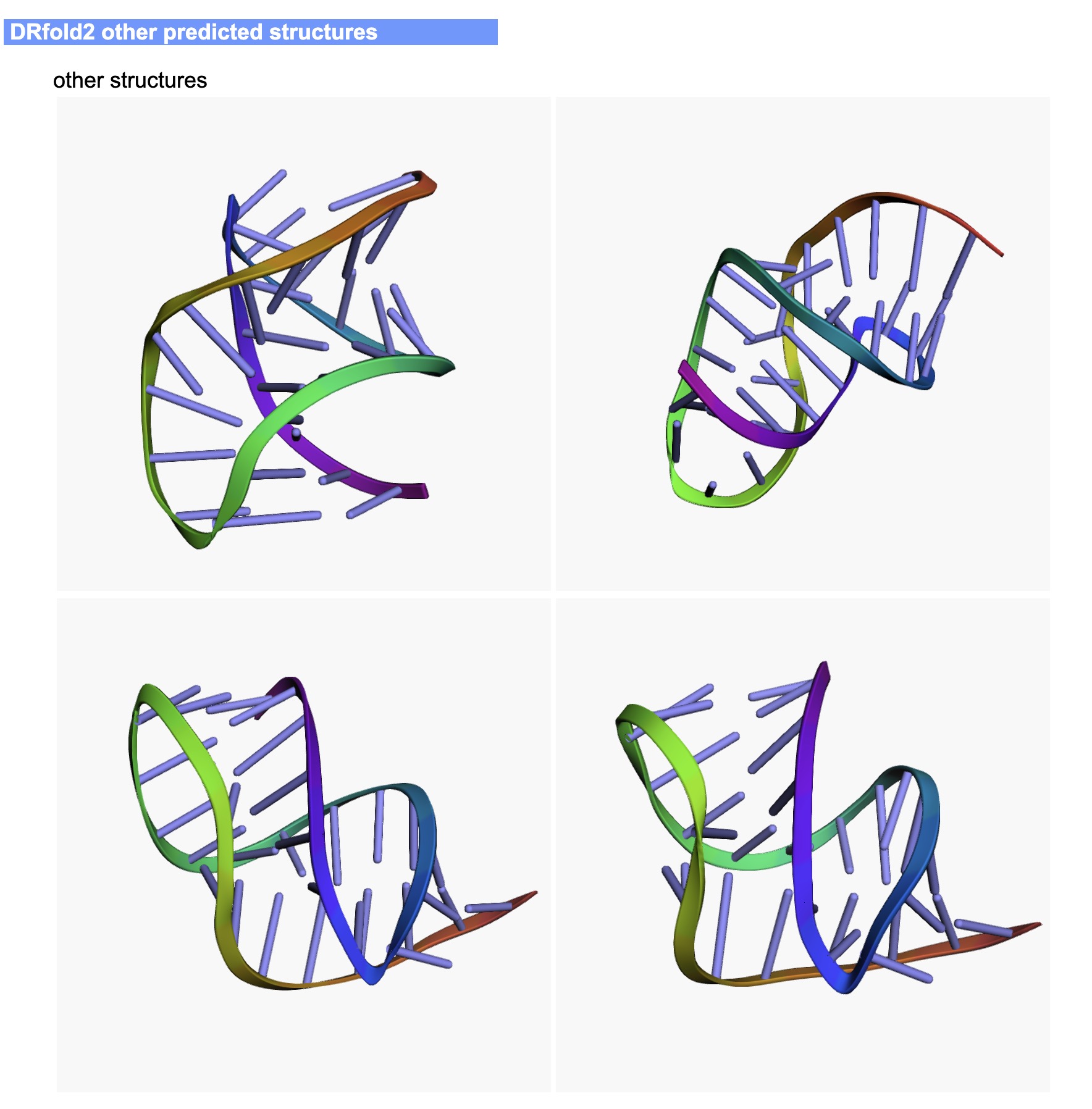
The output of the DRfold2 server include:
- The full-length atomic model
- Predicted geometry maps from geometry models
- Other predicted conformations
An illustrative example of the DRfold2 output can be seen from below (Figs. 4-6):
How to cite DRfold?
- Yang Li, Chenjie Feng, Xi Zhang, Yang Zhang. "Ab initio RNA structure prediction with composite language model and denoised end-to-end learning", submitted.
[back to the DRfold2 server]


 Figure 1. Pipeline of DRfold2.
Figure 1. Pipeline of DRfold2.
 Figure 2. Performance comparisons between DRfold2 and
other RNA structure prediction methods.
Figure 2. Performance comparisons between DRfold2 and
other RNA structure prediction methods.

![]() zhanggroup.org
| +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org
| +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417