
Methods
Starting from a query sequence, DRfold (Deep RNA fold) will use the sequence and its predicted secondary structures as the inputs of the deep learning model. The deep learning model employs 48 transformer blocks
with pair representations for end-to-end RNA coordinates prediction. The pair representations are also used
for inter-nucleotide geometry prediction (e.g., distance and dihedral angle maps). A novel potential that combines
information from end-to-end and geometry predictions is developed to guide the L-BFGS structure optimization to obtain the final
structure.
(see Fig. 1).
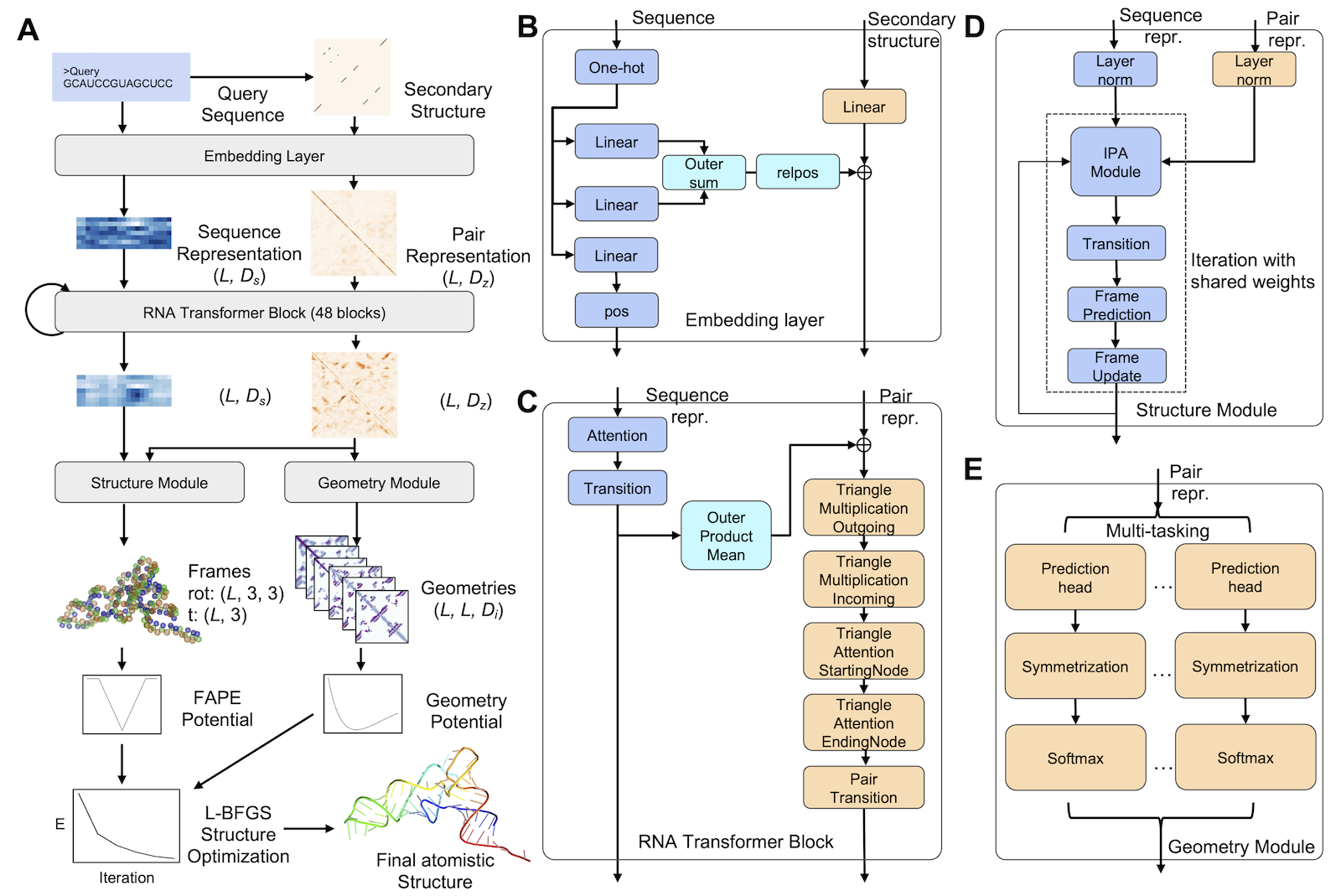 Figure 1. Pipeline of DRfold for deep-learning based ab initio RNA structure prediction.
Figure 1. Pipeline of DRfold for deep-learning based ab initio RNA structure prediction.
Performance
DRfold was tested on a set of 40 recently released targets collected from PDB. The performance of DRfold is significantly better
than existing RNA structure predictors. Fig. 2 compares the performance evaluated by RMSD and TM-score of P atoms.
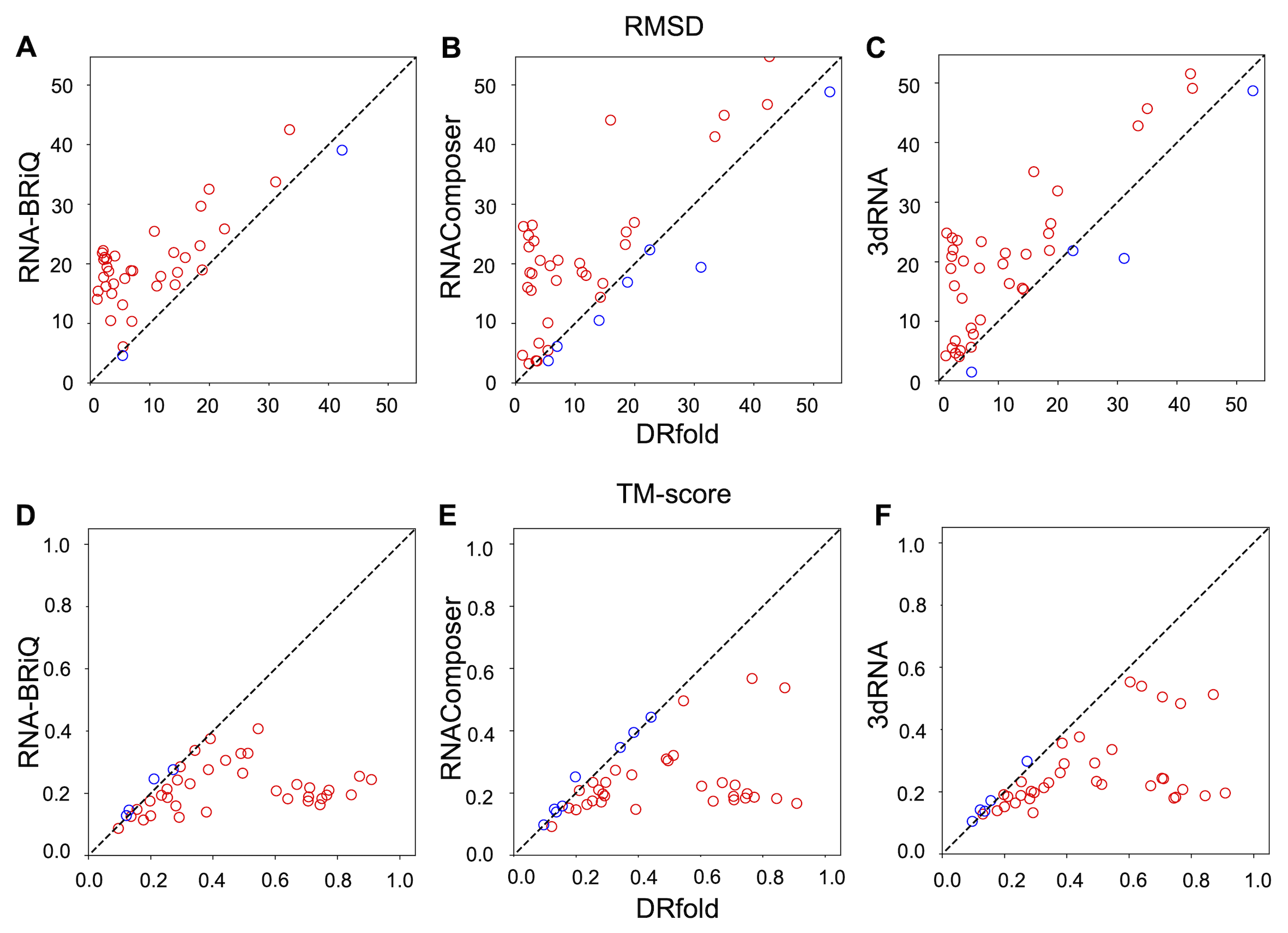 Figure 2. Head-to-head performance comparisons between DRfold and
other RNA structure prediction methods.
Figure 2. Head-to-head performance comparisons between DRfold and
other RNA structure prediction methods.
Server inputs
The user needs to paste the RNA sequence into the input box,
or upload the sequence of the query RNA using the "Choose file" button (see Fig. 3).
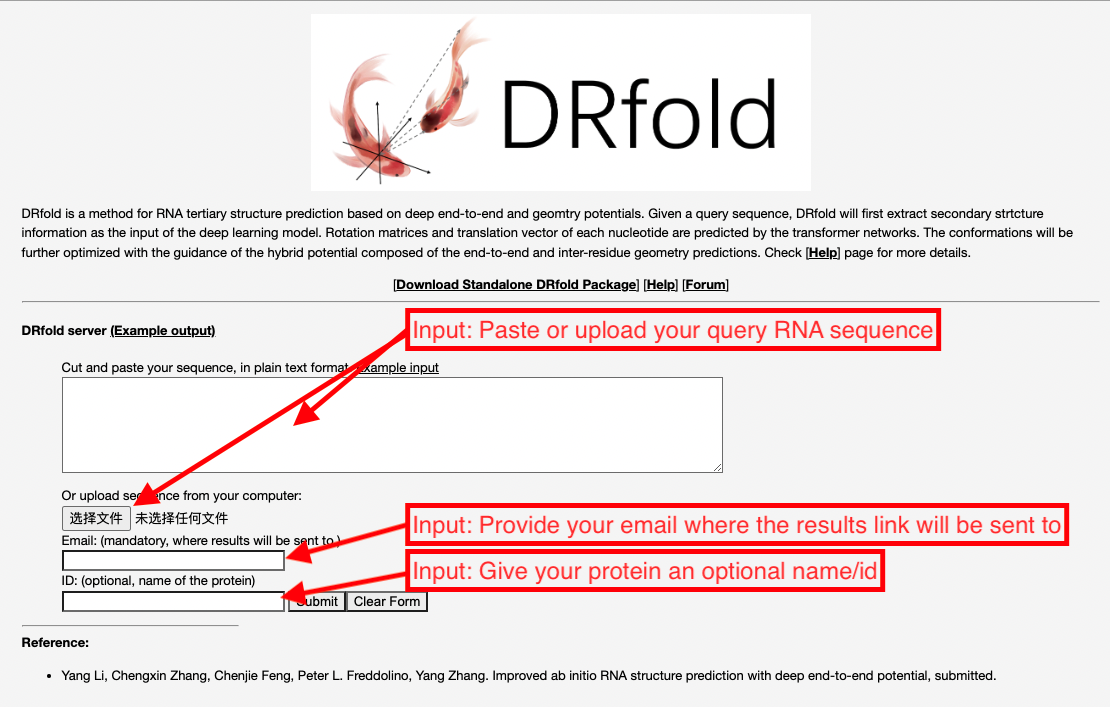
Figure 3. Illustration of input for the DRfold server.
Server outputs
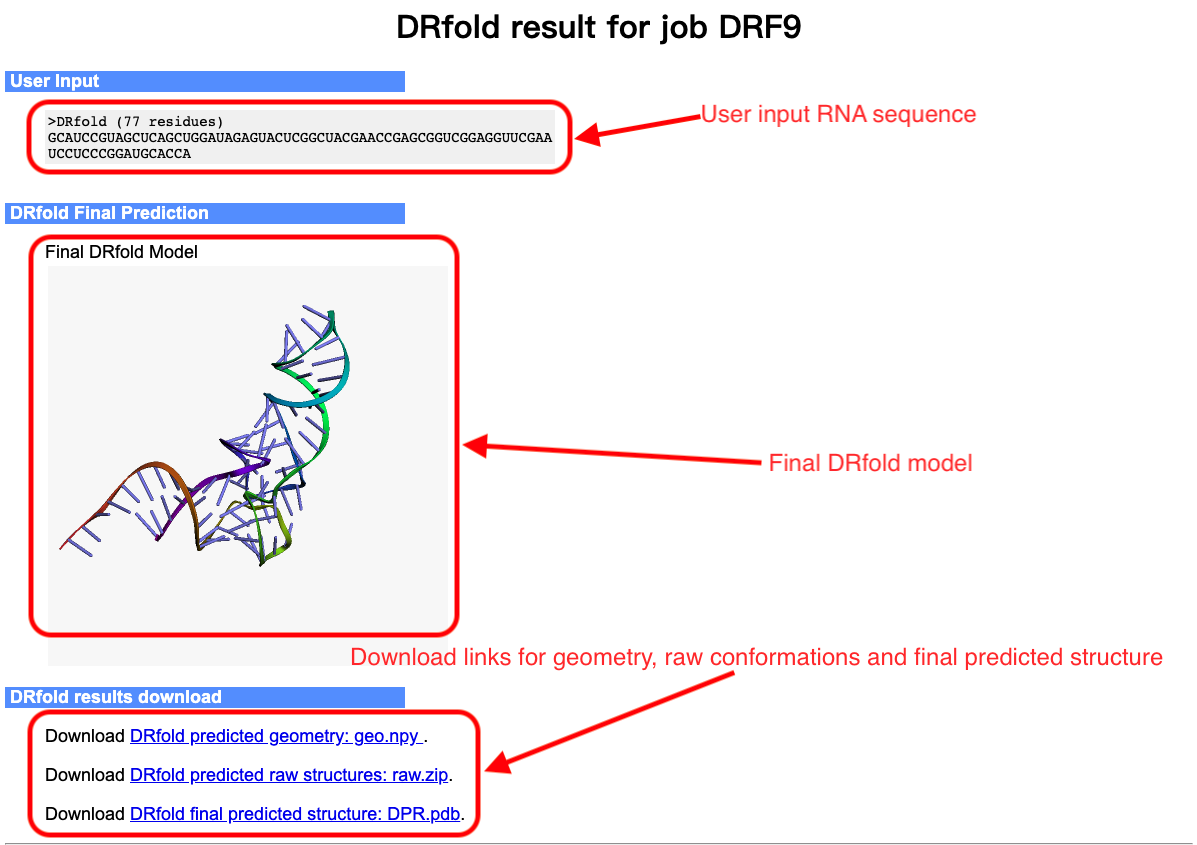
The output of the DRfold server include:
- The full-length atomic model
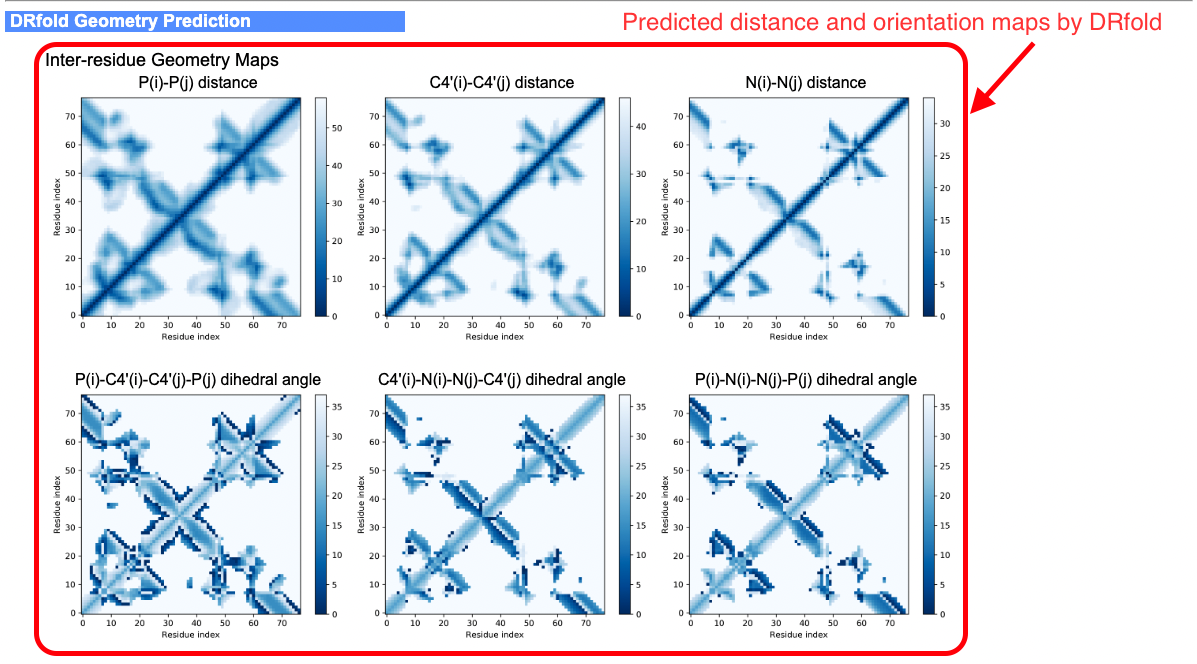
- Predicted geometry maps from geometry models
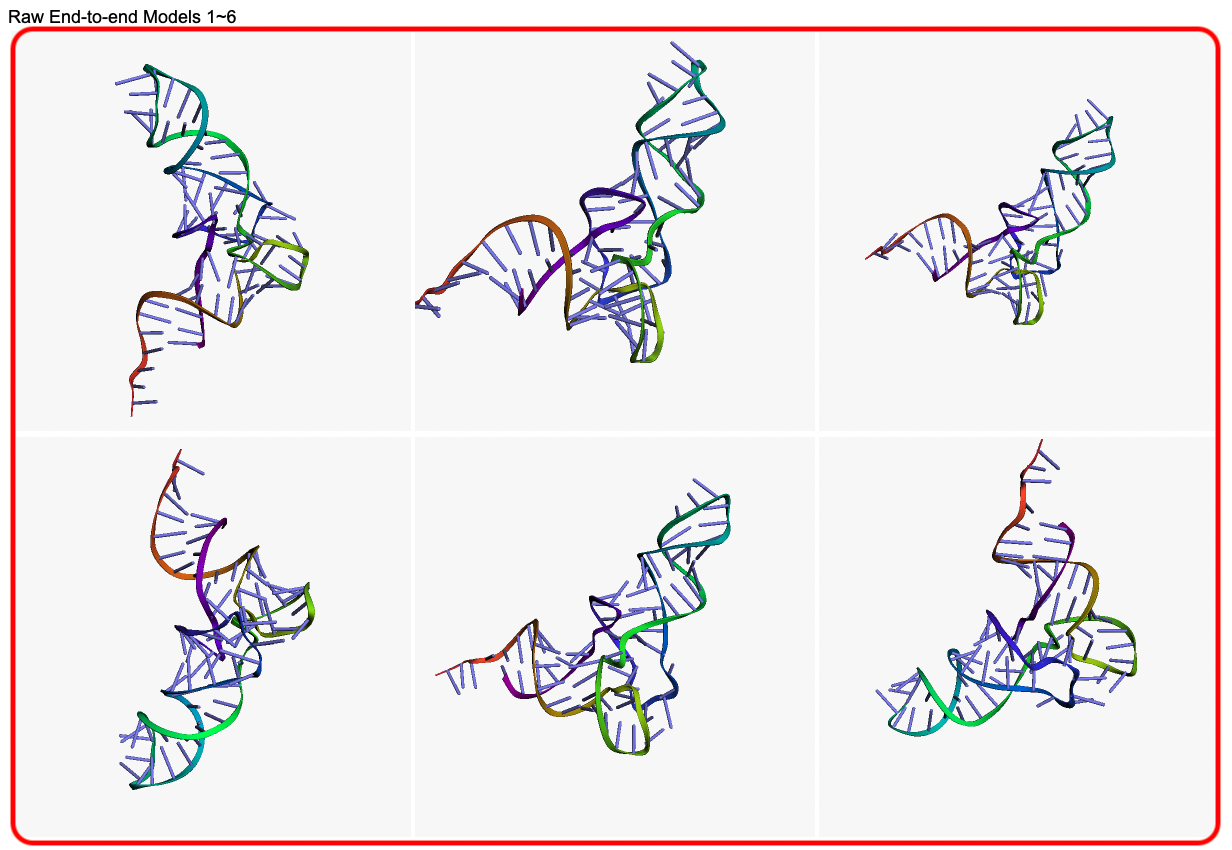
- Predicted raw conformations from end-to-end models
An illustrative example of the DRfold output can be seen from below (Figs. 4-6):
How to cite DRfold?
-
Yang Li, Chengxin Zhang, Chenjie Feng, Robin Pearce, P. Lydia Freddolino, Yang Zhang.
"Integrating end-to-end learning with deep geometrical potentials for ab initio RNA structure prediction." Nature Communications 14, no. 1 (2023): 5745.
[back to the DRfold server]



 Figure 1. Pipeline of DRfold for deep-learning based ab initio RNA structure prediction.
Figure 1. Pipeline of DRfold for deep-learning based ab initio RNA structure prediction.
 Figure 2. Head-to-head performance comparisons between DRfold and
other RNA structure prediction methods.
Figure 2. Head-to-head performance comparisons between DRfold and
other RNA structure prediction methods.

![]() zhanggroup.org
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218