
[
Home]
[
Forum]
[
Example
in Default Mode]
[
Help]
(
Note:
The results presented in this page was generated by the "Benchmark"
mode of I-TASSER-MR, in which all homologous templates with a sequence
identity >30% to the query or detectable by PSI-BLAST with E-value <0.001
have been excluded in the I-TASSER modeling.
The results of the default mode of I-TASSER-MR, which has no homology
filter, can be found
Here")
Sequence Information of T23063RE1(your_protein)
|
SMQEEDTFRELRIFLRNVTHRLAIDKRFRVFTKPVDPDEVPDYVTVIKQPMDLSSVISKIDLHKYLTVKDYLRDIDLICS
NALEYNPDRDPGDRLIRHRACALRDTAYAIIKEELDEDFEQLCEEIQESR
|
The Input Parameters
|
| Unit cell (a,b,c): | (79.6160,79.6160,137.5500) |
| Unit cell (α,β,γ): | (90.0000,90.0000,120.0000)
|
| Space Group: | P 65 2 2
|
| Copy Number: | 1
|
| Lowest Resolution: | 68.949
|
| Highest Resolution: | 1.950
|
| mtz file: | Download
|
Best Models with the lowest R-free
I-TASSER-MR Results of Sequence T23063RE1
I-TASSER
Models | I-TASSER-MR Models |
|---|
| Model1 |
|
- I-TASSER Models:the models predicted by I-TASSER;
- I-TASSER-MR Models: top 5 search models, MR models and refined models ranked by the final Rfreee of the refined models;
- Search Model: top 5 search models edited by progressive model truncation;
- MR Model: top 5 models produced by MR-REX from the corresponding top 5 search models;
- Starting Rwork: the starting Rwork of the refined models;
- Final Rwork: the final Rwork of the refined models;
- Starting Rfree: the starting Rfree of the refined models;
- Final Rfree: the final Rfree of the refined models.
|
I-TASSER Search Models for Sequence T23063RE1
I-TASSER
Models | Distribution of the AVS-score | Download |
|---|
Model1
cscore:0.10 | 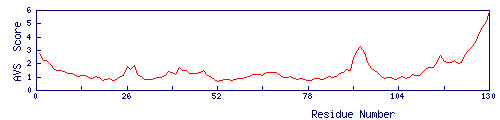 | Download |
|
- I-TASSER Models: the models predicted by I-TASSER;
- Download: the dowload file includes the AVS score file and all the edited search models;
|
Top 10 templates used by I-TASSER for Sequence T23063RE1
| Rank | Template | Aligned Length | Coverage | Z-score | Program | Alignments |
| 1 | 4nxjA | 118 | 0.908 | 17.409 | MUSTER | Download |
| 2 | 5ulc_X | 115 | 0.885 | 16.771 | HHpred_local | Download |
| 3 | 5e74a | 113 | 0.869 | 19.383 | SP3 | Download |
| 4 | 4nxjA | 118 | 0.908 | 7.117 | PROSPECT2 | Download |
| 5 | 4nxjA | 118 | 0.908 | 35.829 | PPA-I | Download |
| 6 | 4uyd_A | 114 | 0.877 | 17.613 | HHpred_global | Download |
| 7 | 4nr9a | 113 | 0.869 | 18.539 | SPARKS2 | Download |
| 8 | 4qnsA | 106 | 0.815 | 16.619 | MUSTER | Download |
| 9 | 4nxj_A | 118 | 0.908 | 16.618 | HHpred_local | Download |
| 10 | 4nr9a | 113 | 0.869 | 19.352 | SP3 | Download |
- Rank of templates: represents the top ten threading templates used by I-TASSER;
- Aligned length: the number of aligned residues;
- Coverage: it represents the coverage of the threading alignment and is equal to the number of aligned residues divided by the length of query protein;
- Z-score: Z-score is the normalized Z-score of the threading alignment;
- Download Alignments: Provides the 3D structure of the aligned regions of the threading templates.
|
Download
References:
- Y. Wang, J. Virtanen, Z. Xue, Y. Zhang.
I-TASSER-MR: automated molecular replacement for proteins without close homologs using iterative
fragment assembly and progressive sequence truncation,
submitted (2017).
- Y. Wang, J. Virtanen, Z. Xue, J. J. G. Tesmer and Y. Zhang. Using iterative fragment assembly and progressive sequence truncation to facilitate phasing and crystal structure determination of distantly related proteins. Acta Cryst. (2016). D72, 616-628


 [Home]
[Forum]
[Example
in Default Mode]
[Help]
[Home]
[Forum]
[Example
in Default Mode]
[Help]
![]() umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218