


HPmod database is a repository for human protein structure and function that automatically predicted by the state-of-the-art algorithm from Yang Zhang Lab. Protein structure models are predicted by D-I-TASSER, and the protein functions, including Gene Ontology (GO), Enzyme Commission (EC), and ligand-binding sites, are predicted by COFACTOR. The database contains 19,512 proteins collected from Uniprot, which can be classified as 12,236 single-domain proteins and 7,276 multi-domain proteins.
As depicted in Figure 1, the overall pipeline of the protein structure prediction designed for human proteome starts from a full-length query sequence. First, the multiple sequence alignment (MSA) for the full-length protein is constructed by DeepMSA methods through iteratively searching genomics and metagenomics sequence databases. The MSA is then used as input for DeepPotential and AttentionPotential to predict geometric restraints, including contact-maps, distances, inter-residue orientations and hydrogen-bond networks. The domain boundaries are predicted by combining the threading template-based method ThreaDom, which is mainly used for Easy targets, with the predicted contact-based method FUpred for Hard targets, resulting in the domain-level sequences. On the one hand, the default D-I-TASSER pipeline is used here to predict domain-level model for each domain-level sequence. The detailed method of D-I-TASSER can be found in D-I-TASSER help page. On the other hand, an L-BFGS system is introduced to construct the full-length structure model for the full-length sequence based on spatial restraints predicted by DeepPotential and AttentionPotential. The individual domain-level models are then assembled into a final full-length structure using the DEMO protocol that uses not only the rough full-length structure but also the templates from PDB database. Starting from the 3D structural model, COFACTOR will thread the query through the BioLiP protein funtion database by local and global structure matches to identify functional sites and homologies. Functional insights, including Gene Ontology (GO), Enzyme Commission (EC), and ligand-binding sites, will be derived from the best functional homology templates.
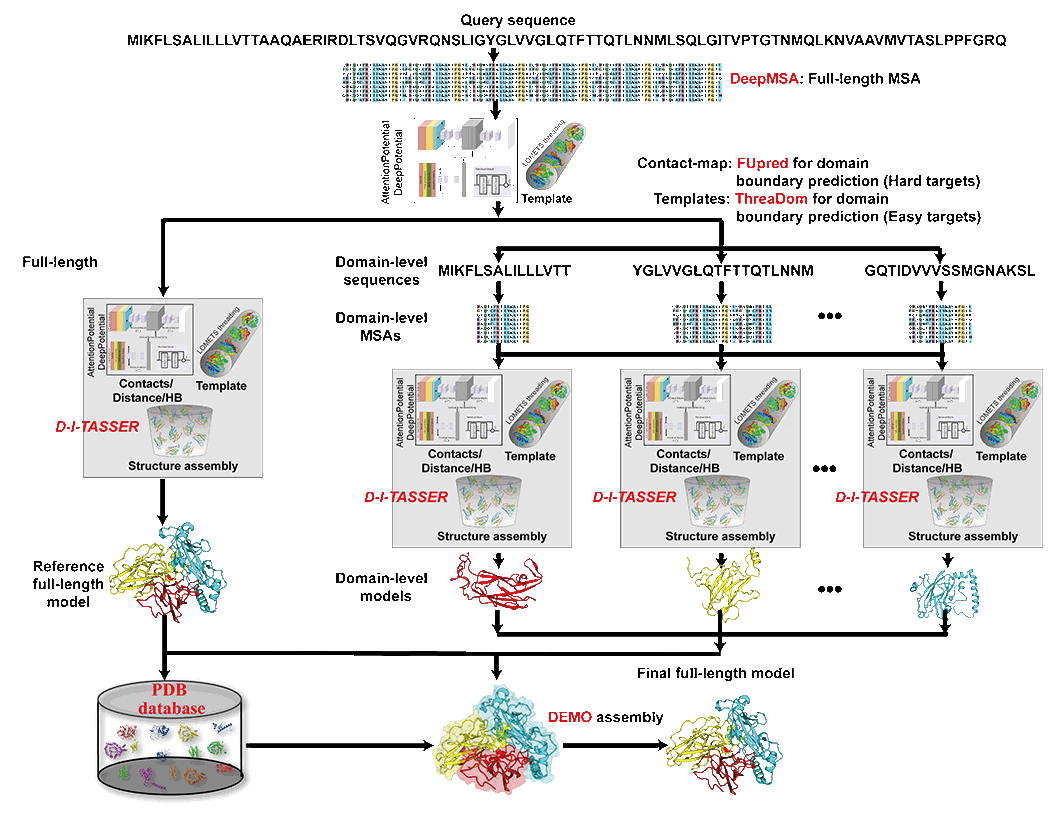 Figure 1. Pipeline of HPmod.
Figure 1. Pipeline of HPmod.
 Figure 2. Summary of protein information in the HPmod output.
Figure 2. Summary of protein information in the HPmod output.
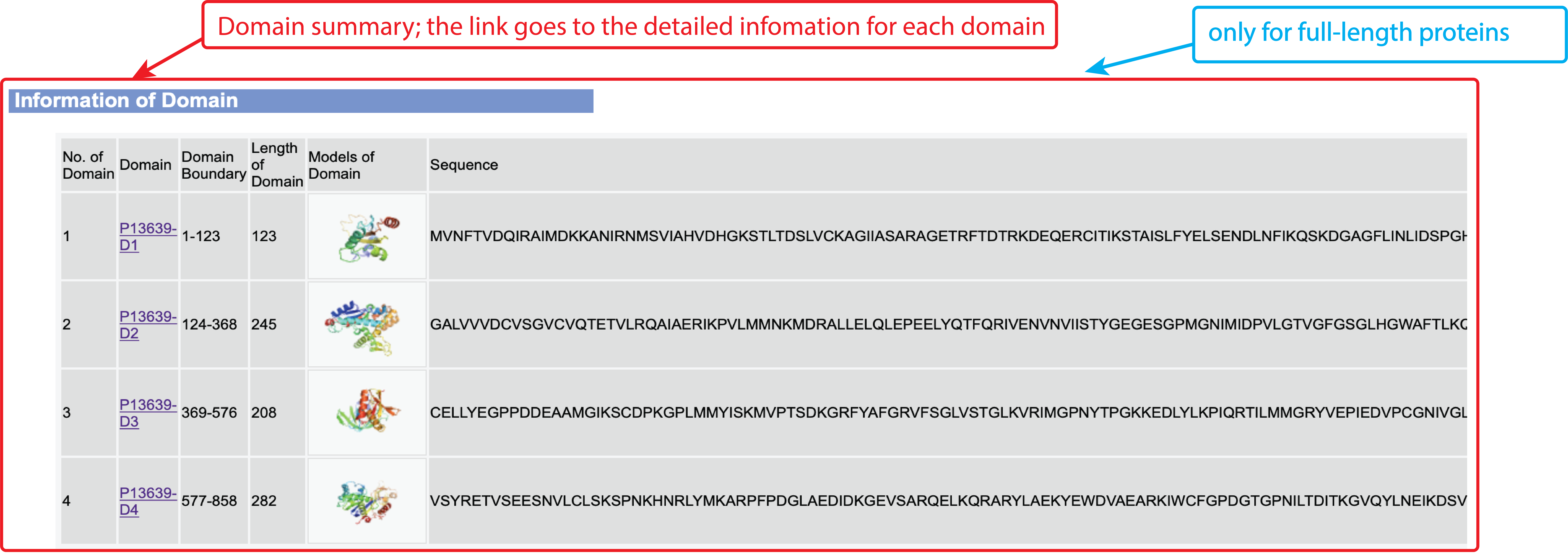 Figure 3. Summary of domain information in the HPmod output.
Figure 3. Summary of domain information in the HPmod output.
 Figure 4. Secondary structure, solvent accessibility, contact map,distance Map, and hydrogen bond networks information in the HPmod output.
Figure 4. Secondary structure, solvent accessibility, contact map,distance Map, and hydrogen bond networks information in the HPmod output.
 Figure 5. Templates, final models, and analog information in HPmod output.
Figure 5. Templates, final models, and analog information in HPmod output.
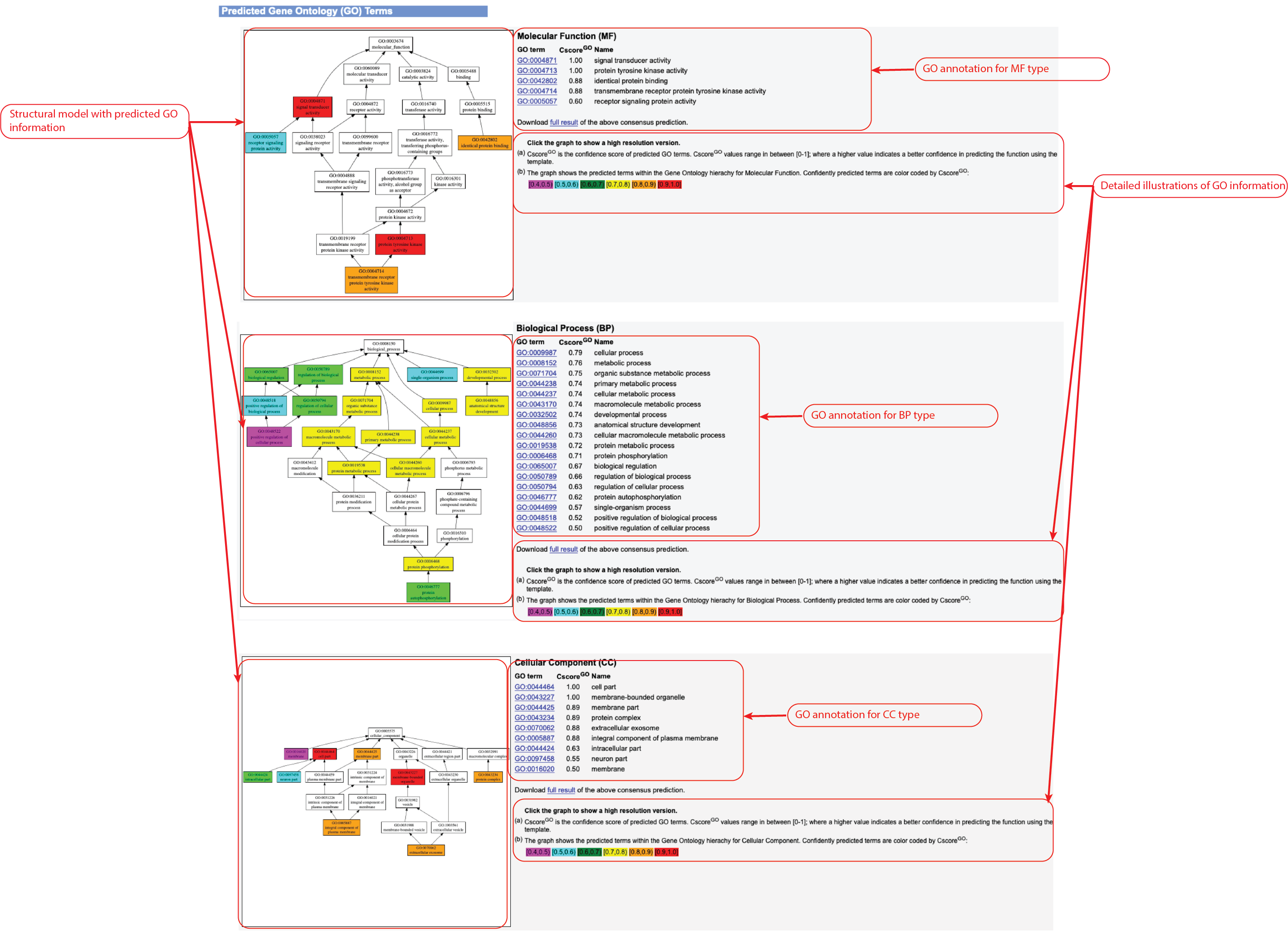 Figure 6. Gene Ontology (GO) Term prediction information in the HPmod output.
Figure 6. Gene Ontology (GO) Term prediction information in the HPmod output.
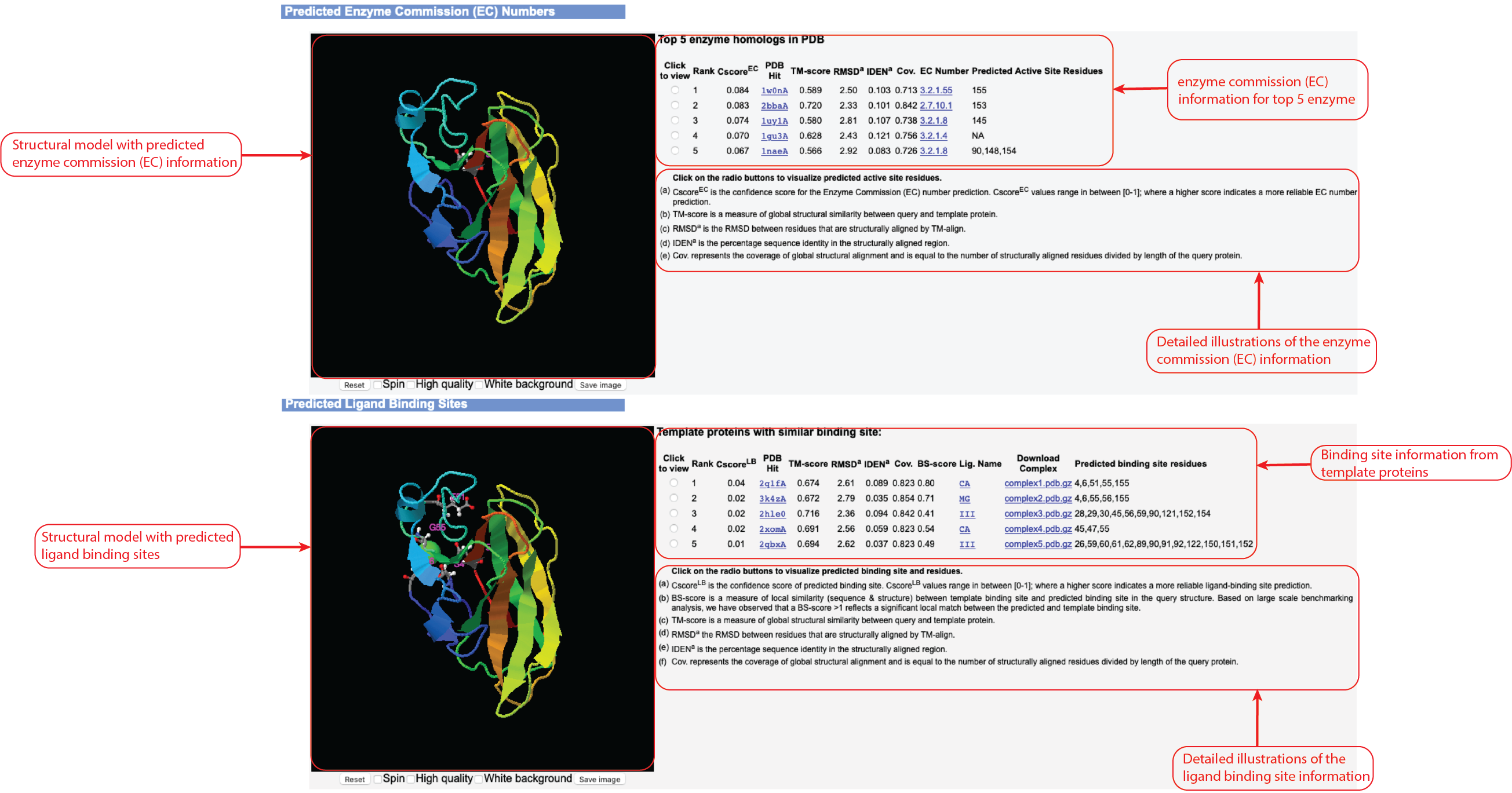 Figure 7. Enzyme Commission (EC) and ligand binding site prediction information in the HPmod output.
Figure 7. Enzyme Commission (EC) and ligand binding site prediction information in the HPmod output.
D-I-TASSER modeling starts from the structure templates identified by LOMETS3 from the PDB library. LOMETS3 is a meta-server threading approach containing multiple threading programs, where each program can generate tens of thousands of templates. D-I-TASSER only uses the templates of the highest significance in the threading alignments, which are measured by the Z-score (the difference between the raw and average scores in the unit of standard deviation). The top 10 templates are the 10 templates selected from the LOMETS3 threading programs. Usually, one (or two) template with the highest Z-score is selected from each threading program, where the threading programs are sorted by the average performance in the large-scale benchmark test experiments.
For each target, D-I-TASSER simulations generate tens of thousands of conformations (called decoys). To select the final models, D-I-TASSER uses the SPICKER program to cluster all the decoys based on pair-wise structure similarity, and report up to five models which correspond to the five largest structure clusters. In Monte Carlo theory, the largest clusters correspond to the states of the largest partition function (or lowest free energy) and therefore have the highest confidence. The confidence of each model is quantitatively measured by eTM-score (see below). Since the top 5 models are ranked by the cluster size, it is possible that the lower-rank models have a higher eTM-score. Although the first model has a higher eTM-score and a better quality in most cases, it is not unusual that the lower-rank models have a better quality than the higher-rank models. If the D-I-TASSER simulations converge, it is possible to have less than 5 clusters generated. This is usually an indication that the models are high quality because of the converged simulations.
After the structure-assembly simulation, D-I-TASSER uses the TM-align program to match the first D-I-TASSER model to all structures in the PDB library. This section reports the top 10 proteins from the PDB which have the closest structural similarity (i.e. the highest TM-score) to the predicted D-I-TASSER model. Due to their structural similarity, these proteins often have similar function to the target. However, users are encouraged to use the function prediction in D-I-TASSER output to obtain the biological function of the target protein, since D-I-TASSER predicts the function using COACH and COFACTOR, which have been extensively trained to derive function from many sequence and structure features, and as a result, these programs have a much higher accuracy than function annotations derived only from the global structure comparison.
Since the experimental structures are unknown for the user input sequence, we have designed an estimated TM-score (eTM-score) to quantitatively estimate the quality of the D-I-TASSER models. The eTM-score is a linear combination of three components: significance of the LOMETS3 threading alignments, satisfaction rate of the predicted contact-maps, the model fitting rate of predicted distance-maps, and the decoy convergence degree of the D-I-TASSER simulations. Based on benchmark testing, the eTM-score had a Pearson correlation coefficient (PCC) of 0.757 with TM-score. As a result of this high correlation, we were able to select a eTM-score cutoff of 0.5, corresponding to an estimated TM-score=0.5, and attain a Matthews correlation coefficient (MCC) on the benchmark dataset of 0.644 and a false discovery rate (FDR) of only 2.71%. Therefore, the D-I-TASSER models with eTM-score > 0.5 are considered to be successfully folded.
eTM-score is designed to quantitatively evaluate the quality of the D-I-TASSER models. It is derived from a linear combination of 4 components, including the significance of LOMETS threading alignments, the satisfaction rate of predicted contact-maps, the model fitting rate of predicted distance-maps, and the decoy convergence degree of D-I-TASSER simulations. A eTM-score of higher value signifies a model of high confidence.
TM-score is a metric for measuring the structural similarity between two structures (see Zhang and Skolnick, Scoring function for automated assessment of protein structure template quality, Proteins, 2004 57: 702-710). The purpose of proposing TM-score is to solve the problem of RMSD which is sensitive to local errors. Because RMSD is an average distance of all residue pairs in two structures, a local error (e.g. a misorientation of the tail) will result in a big RMSD value although the global topology is correct. In TM-score, however, the small distance is weighted stronger than the big distance, which makes the score insensitive to local modeling errors. A TM-score > 0.5 indicates a model of correct topology and a TM-score < 0.17 means a random similarity. These cutoffs are not dependent on the protein length.
TM-score (or RMSD) is a known standard for measuring structural similarity between two structures and is typically used to measure the accuracy of structure modeling when the native structure is known. eTM-score is a metric that was developed for D-I-TASSER to estimate the confidence of modeling. In the case where the native structure is not known, it becomes necessary to use the eTM-score predict the quality of the modeling prediction, i.e. the distance between the predicted model and the native structures.
In a benchmark test set of 797 proteins, we found that eTM-score is highly correlated with TM-score. The correlation coefficient of the eTM-score of the first model with the TM-score to the native structure is 0.757. These data lay the base for the reliable prediction of the TM-score using eTM-score. In the output section, D-I-TASSER reports the eTM-scores of all predicted models for reference.
zhanglab![]() zhanggroup.org
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218