


|
This page contains 3D structural models (Version 3, built on Aug 2014) of 1,026 putative G protein-coupled receptors (GPCRs) in the human genome generated by the GPCR-I-TASSER pipeline. The most recent (Version 4, built on June 2018) is now available as part of the GPCR-EXP database. In GPCR-I-TASSER, the GPCR sequences are first threaded through the GPCR template library to identify muliple structure templates by the LOMETS programs. When close homolgous templates are identified, full-length models will be constructed by the I-TASSER based fragment assembly simulations, assisted by a GPCR and membrane specific force field and spatial restraints collected from mutagenesis experiments in GPCR-RD. In case that homologous templates are not available, an ab initio folding procedure is used to assemble the 7-TM-helix bundle from scratch, followed by the GPCR-I-TASSER fragment reassembly simulations. For multiple domain GPCRs, structural models are built by GPCR-I-TASSER for each domain separately which are then reassembly by the I-TASSER approach. All the models are finally subjected to FG-MD for fragment-guided molecular dynamic simulation refinements. Note:
|
Other GPCR-related resources
|
| HG ID | UniProt ID | Length | C-score | Estimated TM-score |
Estimated RMSD |
Top 5 models |
| HG0400 | B2R9L7 | 389 | -0.38 | 0.66 ± 0.13 | 7.6 ± 4.3 |  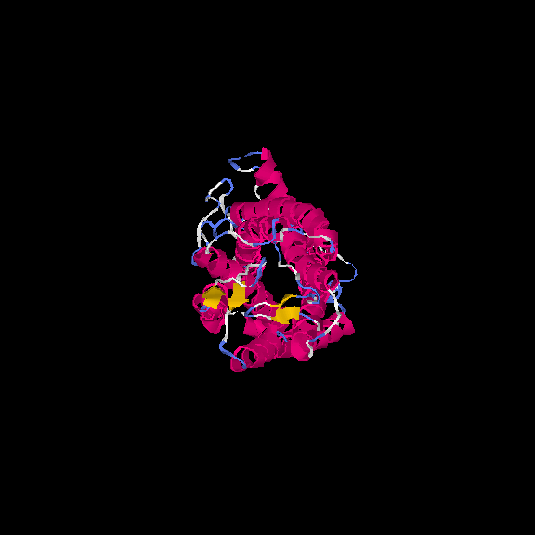 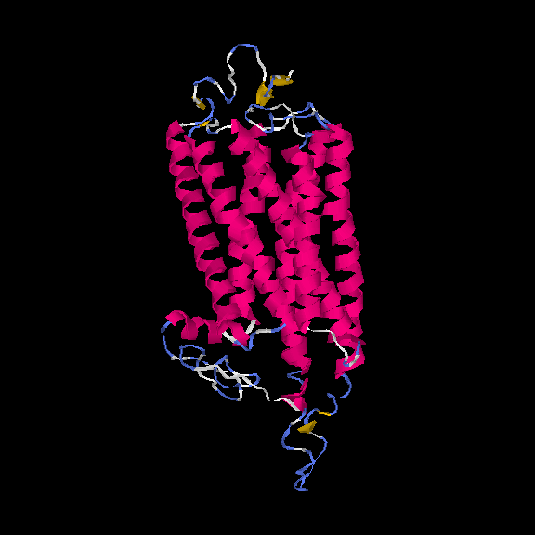 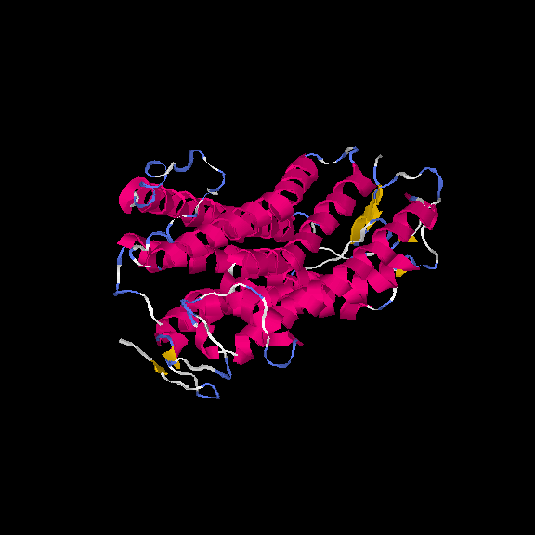 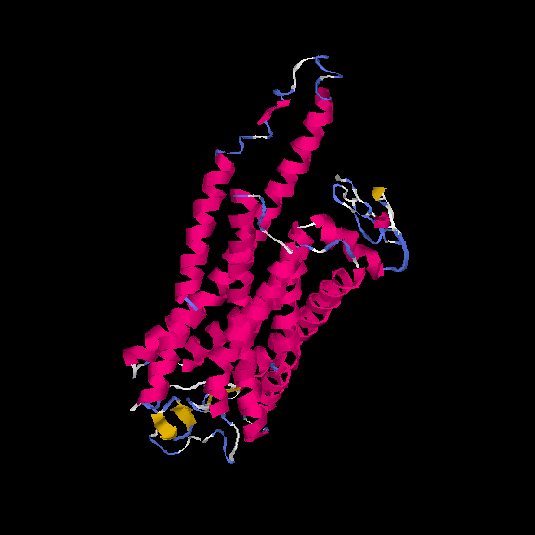 |
| HG0401 | Q9GZP7 | 353 | -1.74 | 0.5 ± 0.15 | 9.99 ± 4.6 |  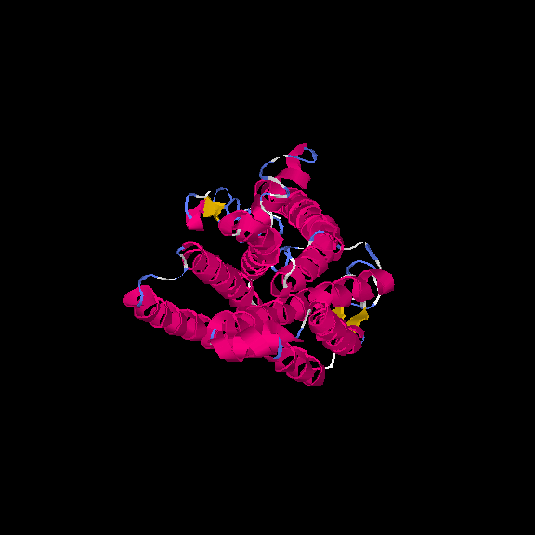 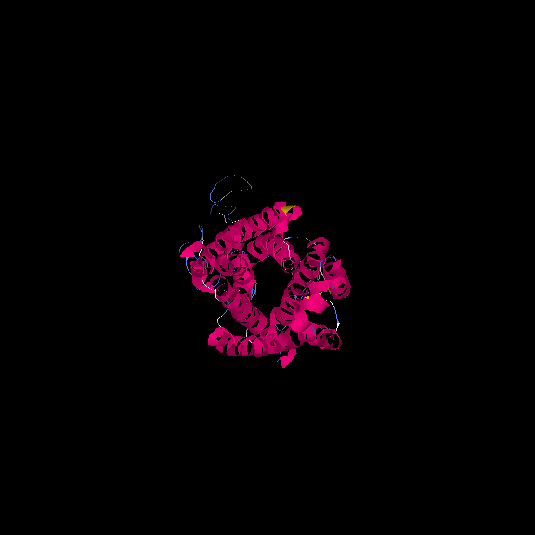 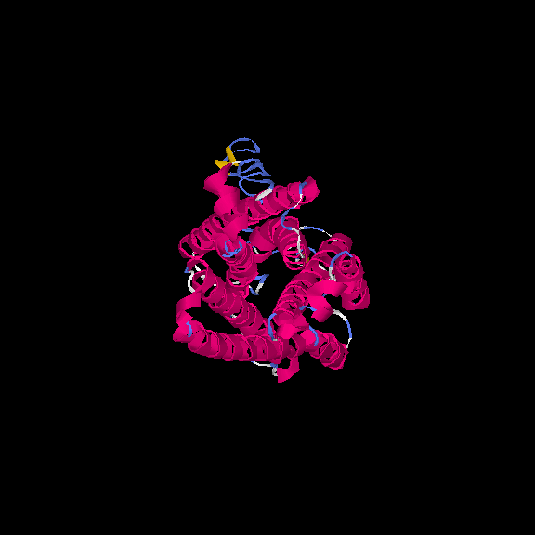 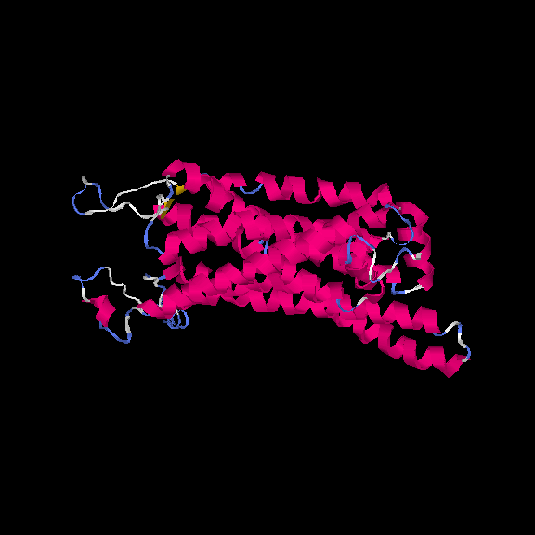 |
| HG0402 | Q8NG98 | 312 | 0.12 | 0.73 ± 0.11 | 6 ± 3.7 | 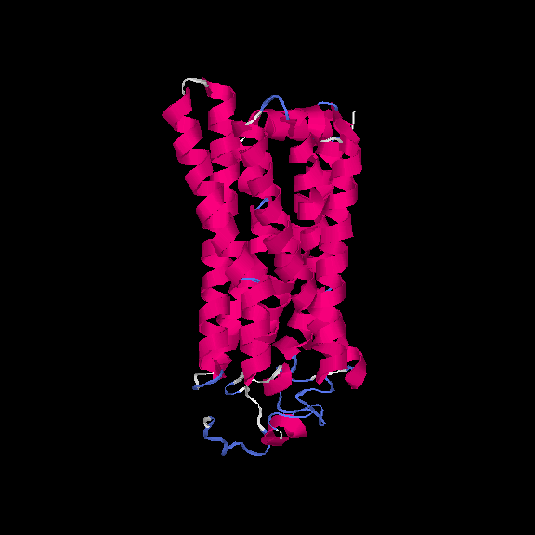 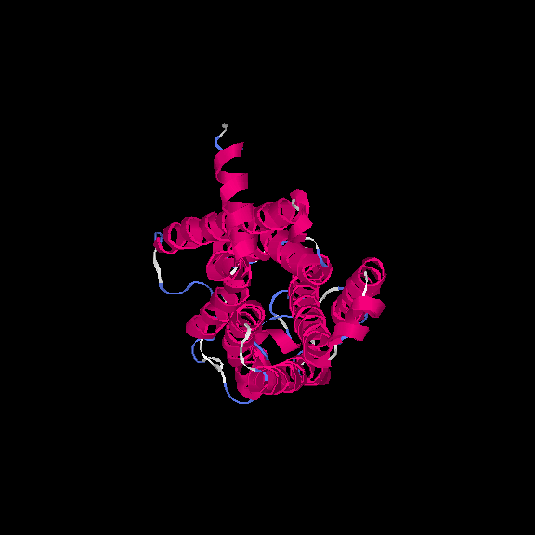 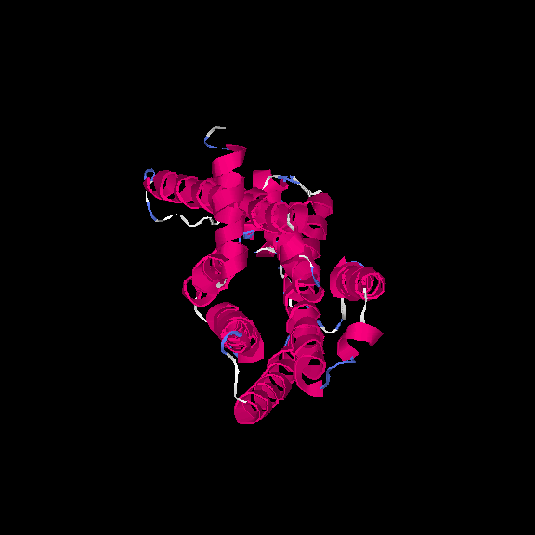 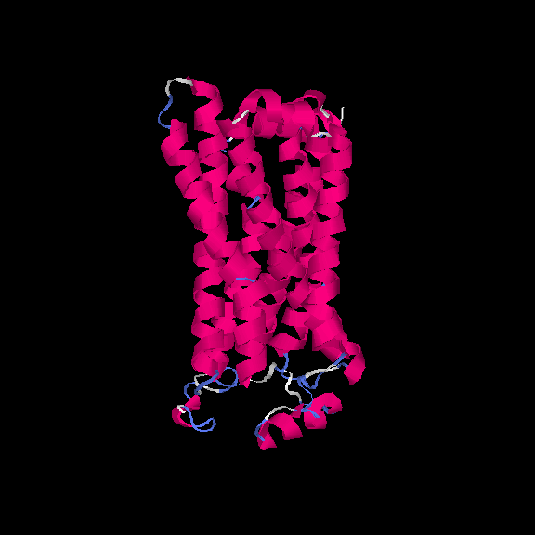 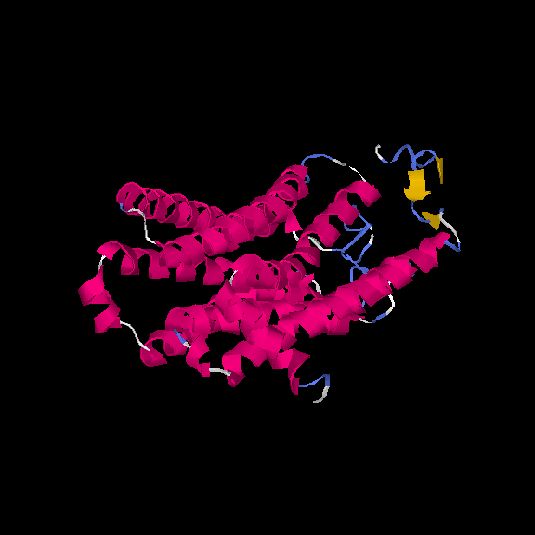 |
| HG0403 | P47901 | 424 | -1.52 | 0.53 ± 0.15 | 9.99 ± 4.6 | 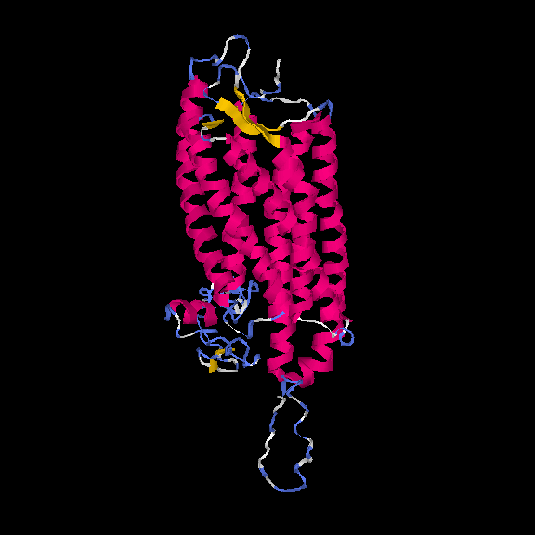  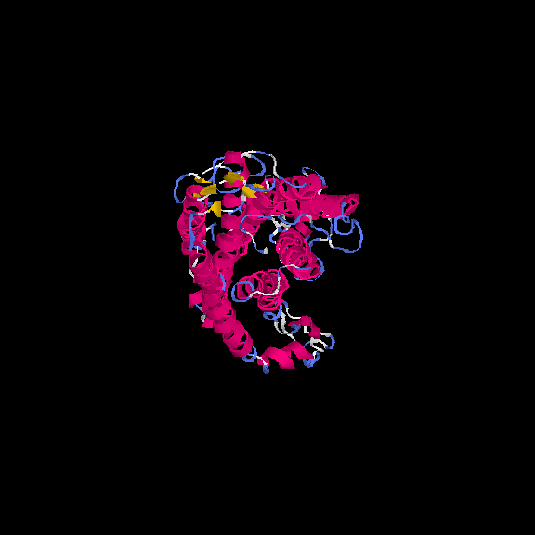  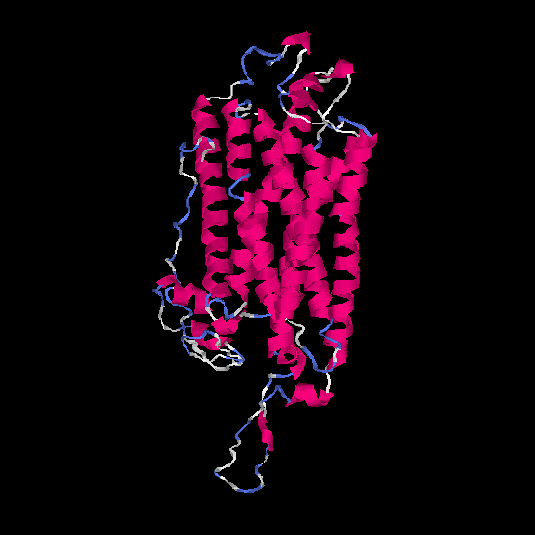 |
| HG0404 | P08172 | 466 | 0.12 | 0.73 ± 0.11 | 6.9 ± 4.1 | 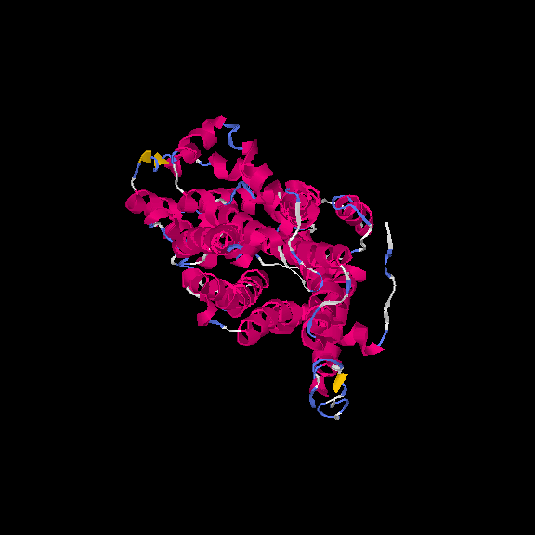 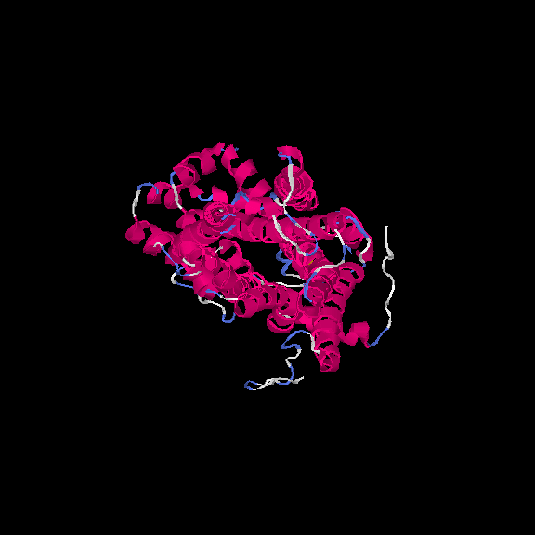 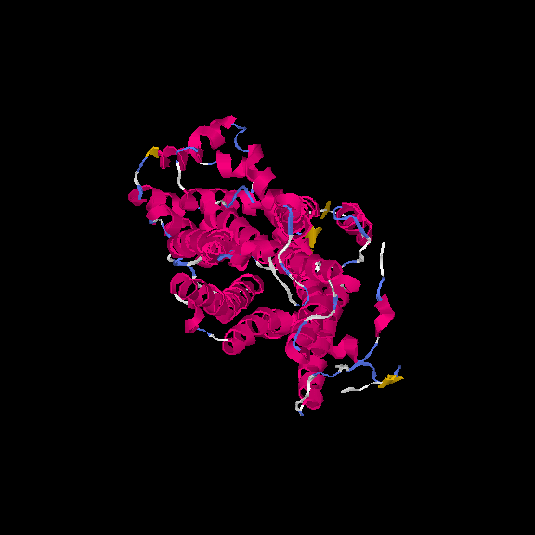 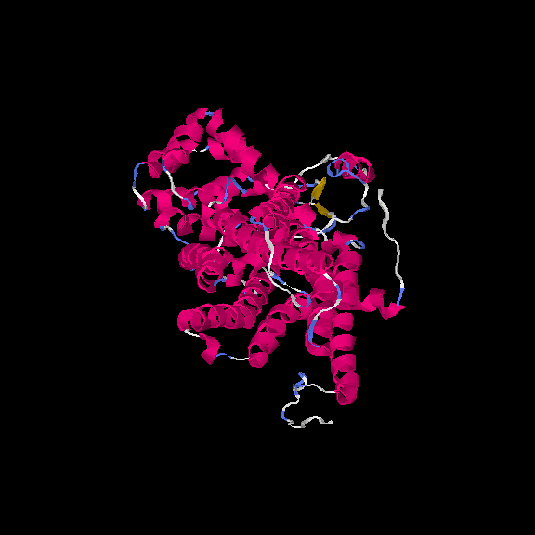  |
| HG0405 | A6NMS3 | 321 | -0.21 | 0.69 ± 0.12 | 6.8 ± 4 | 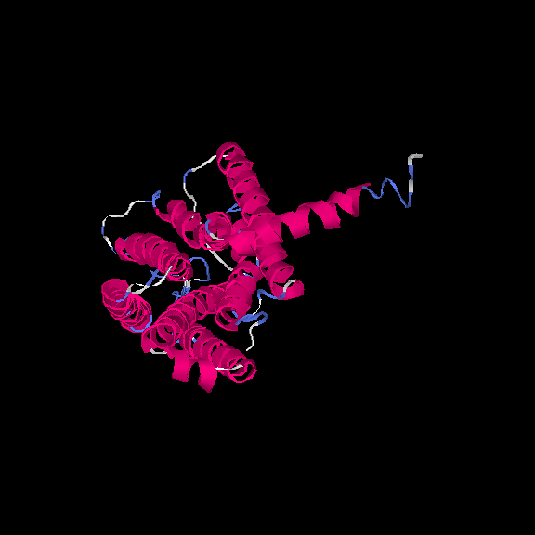 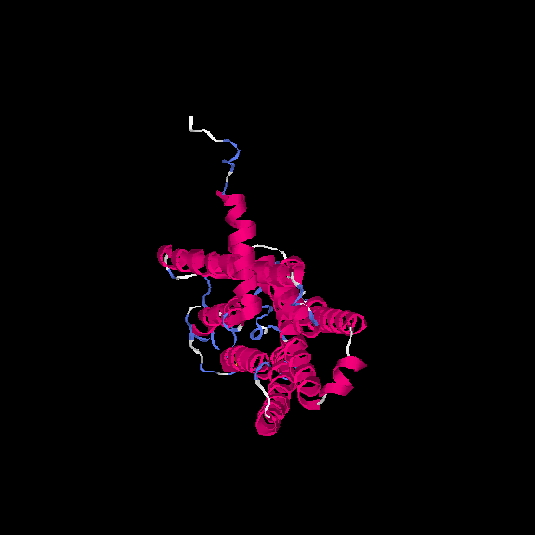 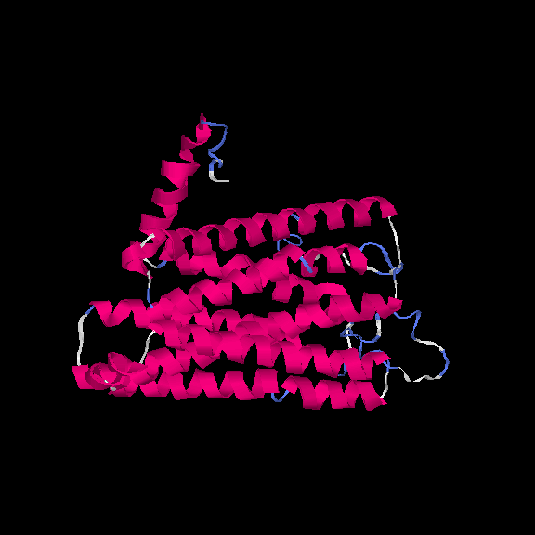 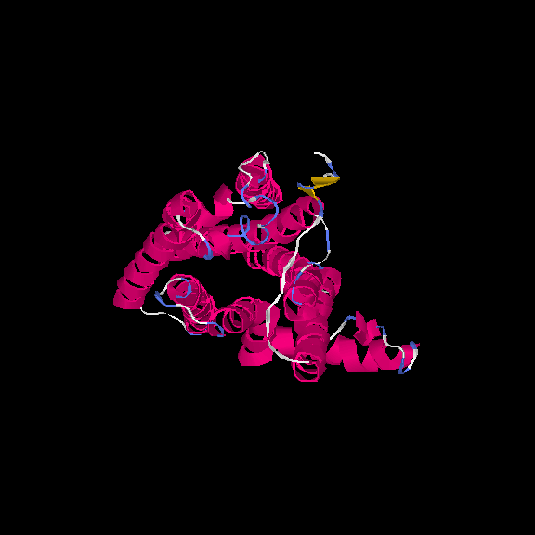 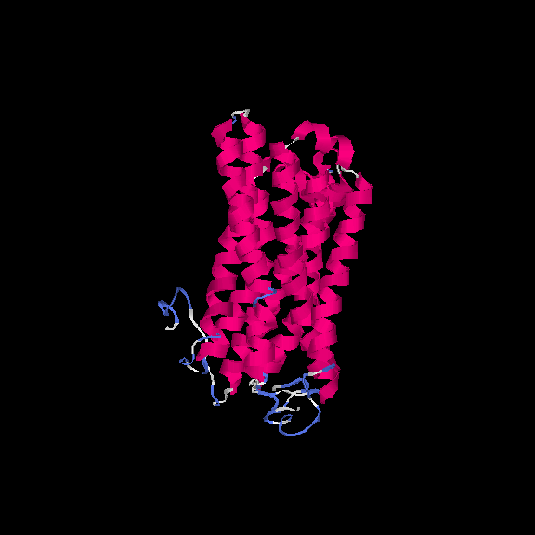 |
| HG0406 | Q8NGW1 | 331 | -0.29 | 0.68 ± 0.12 | 7 ± 4.1 | 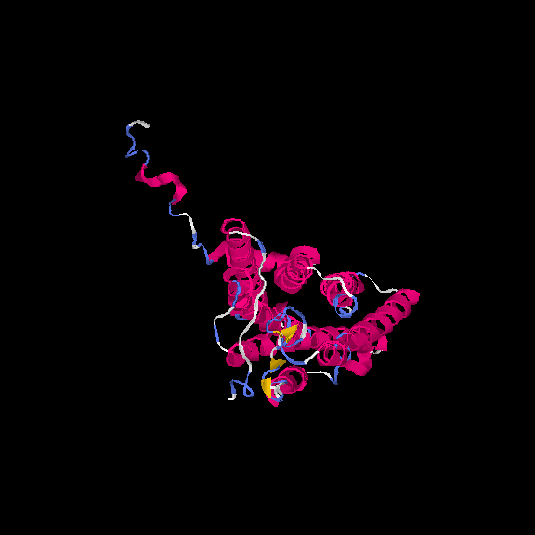 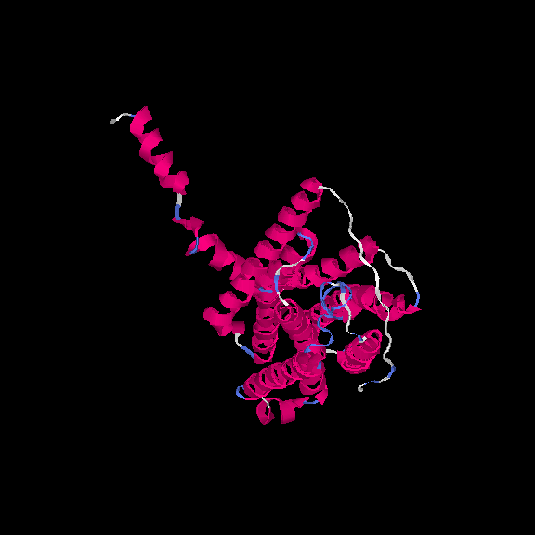 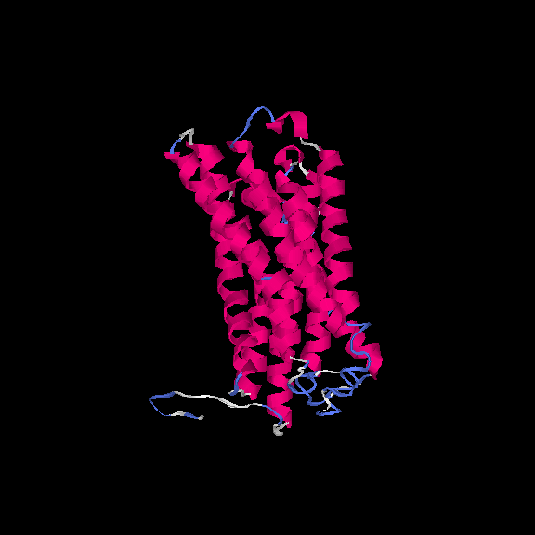 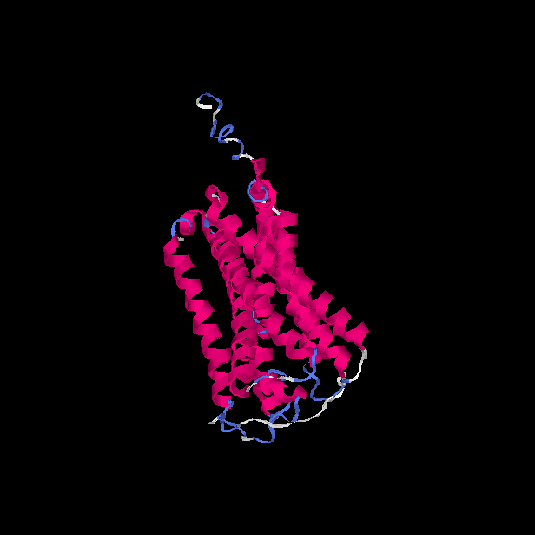 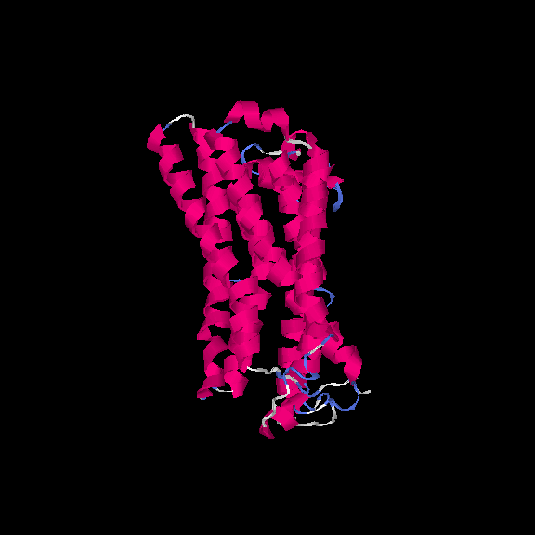 |
| HG0407 | Q1JUL6 | 317 | -0.41 | 0.66 ± 0.13 | 7.2 ± 4.2 |  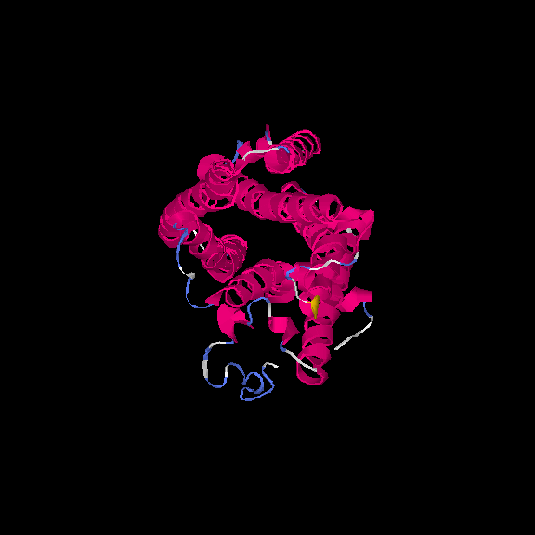 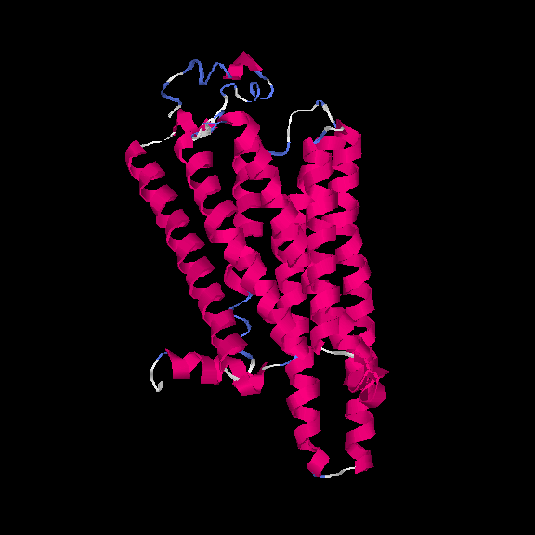 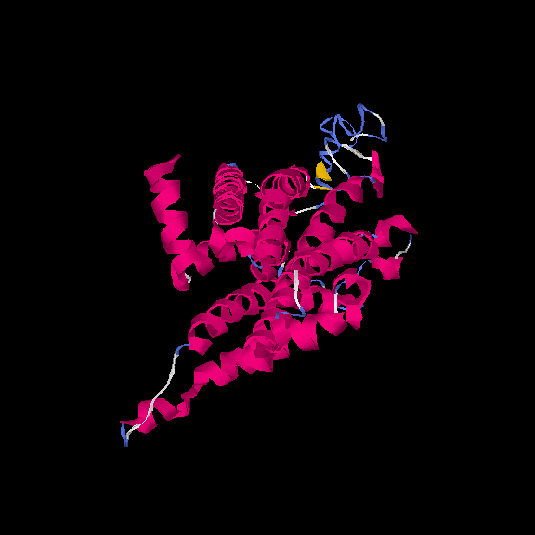 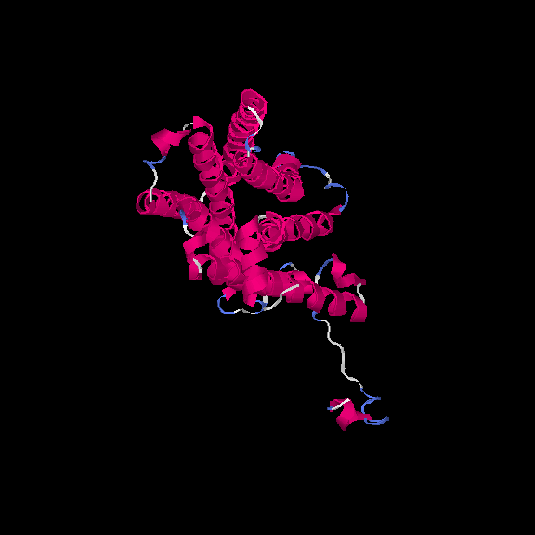 |
| HG0408 | P30542 | 326 | -0.38 | 0.66 ± 0.13 | 7.2 ± 4.2 | 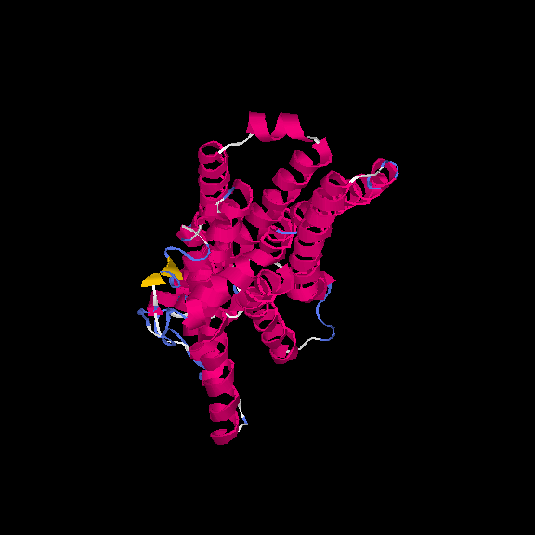 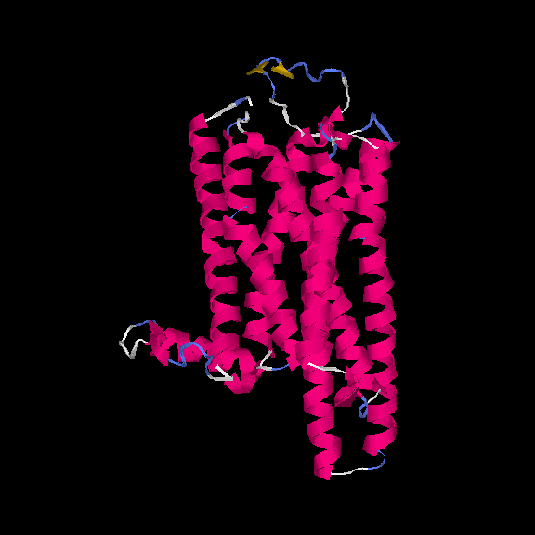    |
| HG0409 | Q8NGC6 | 315 | -0.4 | 0.66 ± 0.13 | 7.2 ± 4.2 |    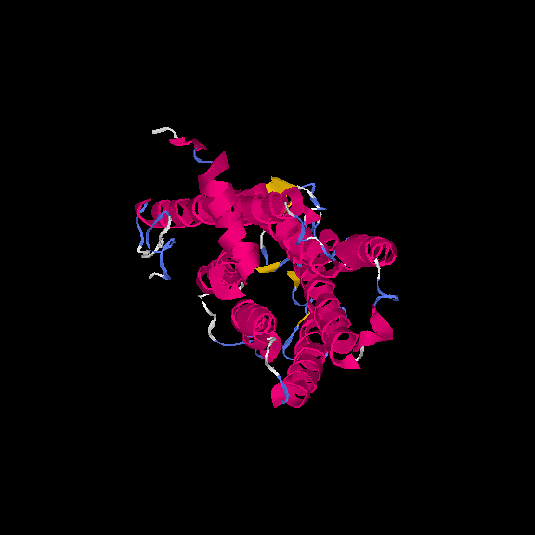  |
| HG0410 | A0N0W7 | 332 | -0.1 | 0.7 ± 0.12 | 6.6 ± 4 | 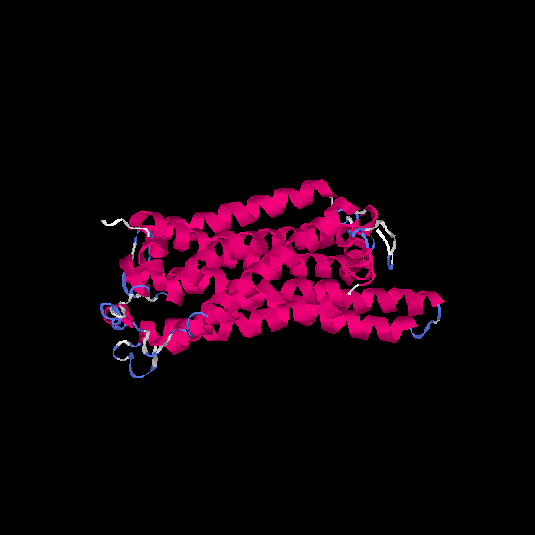 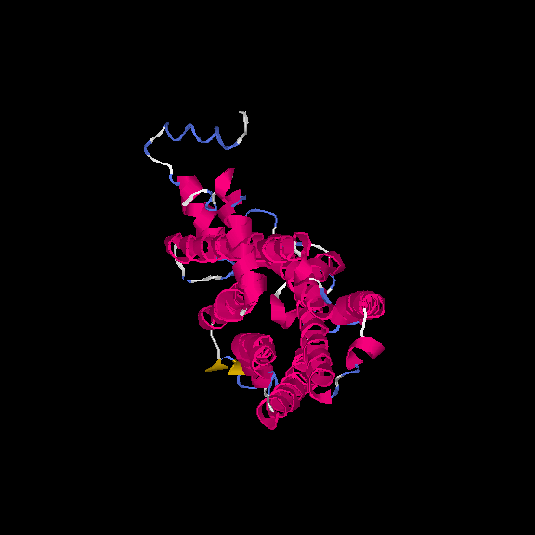 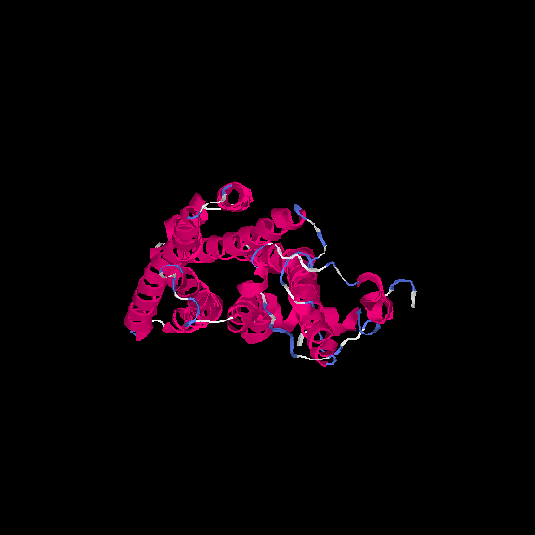   |
zhanglab![]() zhanggroup.org
| +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org
| +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417