

You can:
| Name | Substance-P receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | TACR1 |
| Synonym | NK-1 receptor Tachykinin receptor 1 TAC1R SPR Substance P receptor [ Show all ] |
| Disease | Cough Depression Depression; Anxiety Diabetes Eczema [ Show all ] |
| Length | 407 |
| Amino acid sequence | MDNVLPVDSDLSPNISTNTSEPNQFVQPAWQIVLWAAAYTVIVVTSVVGNVVVMWIILAHKRMRTVTNYFLVNLAFAEASMAAFNTVVNFTYAVHNEWYYGLFYCKFHNFFPIAAVFASIYSMTAVAFDRYMAIIHPLQPRLSATATKVVICVIWVLALLLAFPQGYYSTTETMPSRVVCMIEWPEHPNKIYEKVYHICVTVLIYFLPLLVIGYAYTVVGITLWASEIPGDSSDRYHEQVSAKRKVVKMMIVVVCTFAICWLPFHIFFLLPYINPDLYLKKFIQQVYLAIMWLAMSSTMYNPIIYCCLNDRFRLGFKHAFRCCPFISAGDYEGLEMKSTRYLQTQGSVYKVSRLETTISTVVGAHEEEPEDGPKATPSSLDLTSNCSSRSDSKTMTESFSFSSNVLS |
| UniProt | P25103 |
| Protein Data Bank | 2ksa, 6e59, 6hlo, 2ks9, 6hlp, 6hll, 2ksb |
| GPCR-HGmod model | P25103 |
| 3D structure model | This structure is from PDB ID 2ksa. |
| BioLiP | BL0101802, BL0437915, BL0437914, BL0437913, BL0434896, BL0101803, BL0101801 |
| Therapeutic Target Database | T47094 |
| ChEMBL | CHEMBL249 |
| IUPHAR | 360 |
| DrugBank | BE0000384 |
| Name | CP-99994 |
|---|---|
| Molecular formula | C19H24N2O |
| IUPAC name | (2S,3S)-N-[(2-methoxyphenyl)methyl]-2-phenylpiperidin-3-amine |
| Molecular weight | 296.414 |
| Hydrogen bond acceptor | 3 |
| Hydrogen bond donor | 2 |
| XlogP | 2.9 |
| Synonyms | (2~{S},3~{S})-~{N}-[(2-methoxyphenyl)methyl]-2-phenyl-piperidin-3-amine GTPL2102 (2-Methoxy-benzyl)-(2-phenyl-piperidin-3-yl)-amine CP 99994 Tritiated-(2-Methoxy-benzyl)-(2-phenyl-piperidin-3-yl)-amine [ Show all ] |
| Inchi Key | DTQNEFOKTXXQKV-HKUYNNGSSA-N |
| Inchi ID | InChI=1S/C19H24N2O/c1-22-18-12-6-5-10-16(18)14-21-17-11-7-13-20-19(17)15-8-3-2-4-9-15/h2-6,8-10,12,17,19-21H,7,11,13-14H2,1H3/t17-,19-/m0/s1 |
| PubChem CID | 5311057 |
| ChEMBL | CHEMBL441225 |
| IUPHAR | 2102 |
| BindingDB | 50000041 |
| DrugBank | N/A |
Structure | 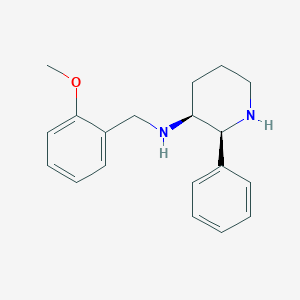 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| ED50 | 0.018 mg.kg-1 | PMID8544174 | ChEMBL |
| IC50 | 0.1698 nM | PMID9733486 | ChEMBL |
| IC50 | 0.17 nM | PMID21413804, PMID9733486 | BindingDB,ChEMBL |
| IC50 | 0.2 nM | PMID7543579 | BindingDB,ChEMBL |
| IC50 | 0.49 nM | PMID9871670 | BindingDB |
| IC50 | 0.49 nM | PMID9871670 | ChEMBL |
| IC50 | 0.5 nM | , PMID8627597, PMID7535362, PMID8709125, Bioorg. Med. Chem. Lett., (1995) 5:13:1345 | BindingDB,ChEMBL |
| IC50 | 0.6 nM | Bioorg. Med. Chem. Lett., (1995) 5:3:209, PMID15481976 | BindingDB,ChEMBL |
| IC50 | 0.6 nM | N/A | BindingDB |
| IC50 | 0.63 nM | PMID17967540 | BindingDB,ChEMBL |
| IC50 | 1.0 nM | , Bioorg. Med. Chem. Lett., (1994) 4:16:1917 | BindingDB,ChEMBL |
| Kd | 1.259 nM | Bioorg. Med. Chem. Lett., (1997) 7:2:203 | ChEMBL |
| Kd | 501187000.0 nM | Bioorg. Med. Chem. Lett., (1997) 7:2:203 | ChEMBL |
| Ki | <10000.0 nM | PMID1282570 | BindingDB,ChEMBL |
| Ki | 0.17 nM | Bioorg. Med. Chem. Lett., (1995) 5:22:2671, PMID1282570 | ChEMBL |
| Ki | 0.17 nM | N/A | BindingDB |
| Ki | 0.199526 - 0.501187 nM | PMID9190866, PMID12206858 | IUPHAR |
| Ki | 0.2512 nM | Bioorg. Med. Chem. Lett., (1995) 5:12:1271, PMID8544174 | ChEMBL |
| Ki | 0.3981 nM | Bioorg. Med. Chem. Lett., (1996) 6:1:13, Bioorg. Med. Chem. Lett., (1996) 6:2:161 | ChEMBL |
| Ki | 0.398107 nM | N/A | BindingDB |
| Ki | 0.48 nM | PMID1282570 | BindingDB,ChEMBL |
| Ki | 0.7943 nM | Bioorg. Med. Chem. Lett., (1995) 5:22:2671 | ChEMBL |
| Ki | 0.794328 nM | N/A | BindingDB |
| Ki | 1.5 nM | , Bioorg. Med. Chem. Lett., (1994) 4:15:1865 | BindingDB,ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417