

You can:
| Name | 5-hydroxytryptamine receptor 2A |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | HTR2A |
| Synonym | 5-HT-2 serotonin receptor 2A serotonin 5HT-2 receptor 5-HT-2A 5-HT2A receptor [ Show all ] |
| Disease | Depression Unspecified Diabetes Erythropoietic porphyria Fibromyalgia [ Show all ] |
| Length | 471 |
| Amino acid sequence | MDILCEENTSLSSTTNSLMQLNDDTRLYSNDFNSGEANTSDAFNWTVDSENRTNLSCEGCLSPSCLSLLHLQEKNWSALLTAVVIILTIAGNILVIMAVSLEKKLQNATNYFLMSLAIADMLLGFLVMPVSMLTILYGYRWPLPSKLCAVWIYLDVLFSTASIMHLCAISLDRYVAIQNPIHHSRFNSRTKAFLKIIAVWTISVGISMPIPVFGLQDDSKVFKEGSCLLADDNFVLIGSFVSFFIPLTIMVITYFLTIKSLQKEATLCVSDLGTRAKLASFSFLPQSSLSSEKLFQRSIHREPGSYTGRRTMQSISNEQKACKVLGIVFFLFVVMWCPFFITNIMAVICKESCNEDVIGALLNVFVWIGYLSSAVNPLVYTLFNKTYRSAFSRYIQCQYKENKKPLQLILVNTIPALAYKSSQLQMGQKKNSKQDAKTTDNDCSMVALGKQHSEEASKDNSDGVNEKVSCV |
| UniProt | P28223 |
| Protein Data Bank | 6a93, 6a94 |
| GPCR-HGmod model | P28223 |
| 3D structure model | This structure is from PDB ID 6a93. |
| BioLiP | BL0441025,BL0441028, BL0441031, BL0441030,BL0441033, BL0441029,BL0441032, BL0441026, BL0441024,BL0441027 |
| Therapeutic Target Database | T32060 |
| ChEMBL | CHEMBL224 |
| IUPHAR | 6 |
| DrugBank | BE0000451 |
| Name | clozapine |
|---|---|
| Molecular formula | C18H19ClN4 |
| IUPAC name | 3-chloro-6-(4-methylpiperazin-1-yl)-11H-benzo[b][1,4]benzodiazepine |
| Molecular weight | 326.828 |
| Hydrogen bond acceptor | 3 |
| Hydrogen bond donor | 1 |
| XlogP | 3.1 |
| Synonyms | NCGC00015254-17 NCGC00022902-08 NINDS_000343 QZUDBNBUXVUHMW-UHFFFAOYSA-N KBio2_000310 [ Show all ] |
| Inchi Key | QZUDBNBUXVUHMW-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C18H19ClN4/c1-22-8-10-23(11-9-22)18-14-4-2-3-5-15(14)20-16-7-6-13(19)12-17(16)21-18/h2-7,12,20H,8-11H2,1H3 |
| PubChem CID | 135398737 |
| ChEMBL | CHEMBL42 |
| IUPHAR | 38 |
| BindingDB | 50001884 |
| DrugBank | DB00363 |
Structure | 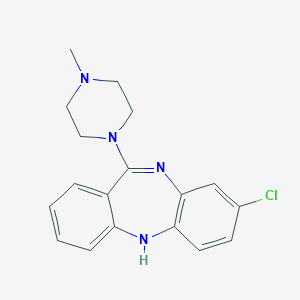 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| N/A | N/A | DrugBank | |
| IC50 | 4.195 nM | DrugMatrix in vitro pharmacology data | ChEMBL |
| IC50 | 12.0 nM | PMID8831770 | BindingDB,ChEMBL |
| Ki | 1.0 - 25.1189 nM | PMID8935801, PMID9732398, PMID15322733, PMID12629531, PMID15102927 | IUPHAR |
| Ki | 1.199 nM | DrugMatrix in vitro pharmacology data | ChEMBL |
| Ki | 2.692 nM | PMID15261283 | ChEMBL |
| Ki | 3.3 nM | PMID14998318 | BindingDB,ChEMBL |
| Ki | 4.0 nM | PMID11170639 | BindingDB,ChEMBL |
| Ki | 4.84 nM | PMID22268448 | BindingDB,ChEMBL |
| Ki | 5.0 nM | PMID8831770 | BindingDB,ChEMBL |
| Ki | 5.012 nM | PMID17880057 | ChEMBL |
| Ki | 5.35 nM | PMID26227779 | ChEMBL |
| Ki | 5.4 nM | PMID26227779 | BindingDB |
| Ki | 6.3 nM | PMID15771415 | BindingDB,ChEMBL |
| Ki | 7.3 nM | PMID24365159 | ChEMBL |
| Ki | 7.413 nM | PMID24365159 | ChEMBL |
| Ki | 7.8 nM | PMID14695828 | BindingDB,ChEMBL |
| Ki | 8.57 nM | Med Chem Res, (2013) 22:2:520 | ChEMBL |
| Ki | 8.9 nM | PMID18595716 | BindingDB,ChEMBL |
| Ki | 9.1 nM | PMID23301527, PMID14741248 | BindingDB,ChEMBL |
| Ki | 9.12 nM | PMID18783204, PMID23301527, PMID24316025, PMID19796944, PMID17588750, PMID14741248, MedChemComm, (2011) 2:12:1194 | BindingDB,ChEMBL |
| Ki | 9.6 nM | , PMID24559051, None | BindingDB,ChEMBL |
| Ki | 10.0 nM | PMID25343529 | BindingDB |
| Ki | 10.0 nM | MedChemComm, (2012) 3:5:580, PMID25343529 | ChEMBL |
| Ki | 15.0 nM | PMID27261181, MedChemComm, (2015) 6:4:601 | BindingDB,ChEMBL |
| Ki | 18.0 nM | PMID25695425 | BindingDB,ChEMBL |
| Ki | 36.0 nM | PMID23301527 | BindingDB,ChEMBL |
| Ki | 36.31 nM | PMID23301527 | ChEMBL |
| Ratio | 1.21 - | PMID11784139 | ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417