| Name | Rhodopsin |
|---|
| Species | Bos taurus (Bovine) |
|---|
| Gene | RHO |
|---|
| Synonym | N/A |
|---|
| Disease | N/A for non-human GPCRs |
|---|
| Length | 348 |
|---|
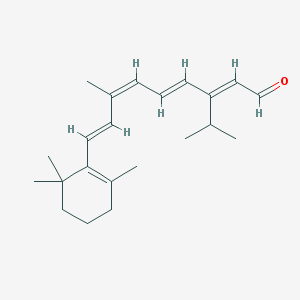
| Amino acid sequence | MNGTEGPNFYVPFSNKTGVVRSPFEAPQYYLAEPWQFSMLAAYMFLLIMLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGYFVFGPTGCNLEGFFATLGGEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVAFTWVMALACAAPPLVGWSRYIPEGMQCSCGIDYYTPHEETNNESFVIYMFVVHFIIPLIVIFFCYGQLVFTVKEAAAQQQESATTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGSDFGPIFMTIPAFFAKTSAVYNPVIYIMMNKQFRNCMVTTLCCGKNPLGDDEASTTVSKTETSQVAPA |
|---|
| UniProt | P02699 |
|---|
| Protein Data Bank | 2i35, 2j4y, 2ped, 3c9l, 3c9m, 2hpy, 2g87, 1u19, 1ln6, 1l9h, 1jfp, 1hzx, 1gzm, 1f88, 2x72, 3dqb, 3oax, 3pqr, 6fkd, 6fkc, 6fkb, 6fka, 6fk9, 6fk8, 6fk7, 6fk6, 5wkt, 5te5, 5en0, 5dys, 4x1h, 4pxf, 4j4q, 4a4m, 3pxo, 6fuf |
|---|
| GPCR-HGmod model | N/A |
|---|
| 3D structure model | This structure is from PDB ID 2i35.
|
|---|
| BioLiP | BL0058001,BL0058005, BL0144059,BL0144060,BL0144061,, BL0144063, BL0149211, BL0186942,BL0186949, BL0186943,BL0186944,BL0186945,, BL0186947,BL0186953, BL0186948,BL0186954, BL0193496, BL0193497, BL0144058, BL0144057, BL0125584, BL0091202,BL0091203,BL0091204,, BL0091206,BL0091210, BL0095323,BL0095324,BL0095325,, BL0095326,BL0095330, BL0096347, BL0099220,BL0099221, BL0107380,BL0107381,BL0107382,, BL0107384,BL0107388, BL0125583, BL0193498, BL0194274, BL0219382, BL0398941, BL0409985, BL0409986, BL0409987, BL0409988, BL0409989, BL0409990, BL0409991, BL0409992, BL0372701, BL0354047, BL0354046, BL0219383, BL0259697, BL0259698,BL0259699,BL0259700,, BL0259702, BL0291136, BL0291137, BL0329157, BL0329158,BL0329159, BL0354035, BL0428638, BL0057997,BL0057998,BL0057999,, BL0032596, BL0398940, BL0012775,BL0012777, BL0012776,BL0012778, BL0018454,BL0018461, BL0018455,BL0018456,BL0018457,, BL0018459,BL0018467, BL0021853,BL0021854,BL0021855,, BL0021857,BL0021861, BL0025799, BL0031623,BL0031624,BL0031625,, BL0031627,BL0031631 |
|---|
| Therapeutic Target Database | N/A |
|---|
| ChEMBL | CHEMBL5739 |
|---|
| IUPHAR | N/A |
|---|
| DrugBank | N/A |
|---|


![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417