

You can:
| Name | P2Y purinoceptor 1 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | P2RY1 |
| Synonym | ATP receptor Purinergic receptor P2Y1 purinergic receptor P2Y Purinergic receptor platelet ADP receptor [ Show all ] |
| Disease | Thrombosis |
| Length | 373 |
| Amino acid sequence | MTEVLWPAVPNGTDAAFLAGPGSSWGNSTVASTAAVSSSFKCALTKTGFQFYYLPAVYILVFIIGFLGNSVAIWMFVFHMKPWSGISVYMFNLALADFLYVLTLPALIFYYFNKTDWIFGDAMCKLQRFIFHVNLYGSILFLTCISAHRYSGVVYPLKSLGRLKKKNAICISVLVWLIVVVAISPILFYSGTGVRKNKTITCYDTTSDEYLRSYFIYSMCTTVAMFCVPLVLILGCYGLIVRALIYKDLDNSPLRRKSIYLVIIVLTVFAVSYIPFHVMKTMNLRARLDFQTPAMCAFNDRVYATYQVTRGLASLNSCVDPILYFLAGDTFRRRLSRATRKASRRSEANLQSKSEDMTLNILPEFKQNGDTSL |
| UniProt | P47900 |
| Protein Data Bank | 4xnv, 4xnw |
| GPCR-HGmod model | P47900 |
| 3D structure model | This structure is from PDB ID 4xnv. |
| BioLiP | BL0311594,BL0311596, BL0311593, BL0311590,BL0311591,BL0311592, BL0311589, BL0311588, BL0311595,BL0311597 |
| Therapeutic Target Database | T67818 |
| ChEMBL | CHEMBL4315 |
| IUPHAR | 323 |
| DrugBank | N/A |
| Name | 2-MeSADP |
|---|---|
| Molecular formula | C11H17N5O10P2S |
| IUPAC name | [(2R,3S,4R,5R)-5-(6-amino-2-methylsulfanylpurin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl phosphono hydrogen phosphate |
| Molecular weight | 473.29 |
| Hydrogen bond acceptor | 15 |
| Hydrogen bond donor | 6 |
| XlogP | -3.8 |
| Synonyms | 2-methylthio-ADP AKOS027379362 methylthio-ADP 2-(Methylthio)-Adenosinediphosphate 6AD [ Show all ] |
| Inchi Key | WLMZTKAZJUWXCB-KQYNXXCUSA-N |
| Inchi ID | InChI=1S/C11H17N5O10P2S/c1-29-11-14-8(12)5-9(15-11)16(3-13-5)10-7(18)6(17)4(25-10)2-24-28(22,23)26-27(19,20)21/h3-4,6-7,10,17-18H,2H2,1H3,(H,22,23)(H2,12,14,15)(H2,19,20,21)/t4-,6-,7-,10-/m1/s1 |
| PubChem CID | 121990 |
| ChEMBL | CHEMBL435402 |
| IUPHAR | 1710, 1763 |
| BindingDB | 50118242 |
| DrugBank | N/A |
Structure | 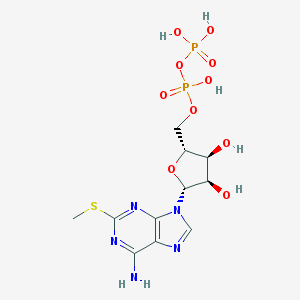 |
| Lipinski's druglikeness | This ligand has more than 5 hydrogen bond donor. This ligand has more than 10 hydrogen bond acceptor. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| EC50 | <10000.0 nM | PMID15481977 | BindingDB,ChEMBL |
| EC50 | <100000.0 nM | PMID15481977 | BindingDB,ChEMBL |
| EC50 | 1.27 nM | PMID15481977 | ChEMBL |
| EC50 | 1.3 nM | PMID15481977 | BindingDB |
| EC50 | 1.58 nM | PMID15481977 | ChEMBL |
| EC50 | 1.6 nM | PMID15481977 | BindingDB |
| EC50 | 1.88 nM | PMID9554879 | ChEMBL |
| EC50 | 1.9 nM | PMID9554879 | BindingDB |
| EC50 | 1.94 nM | PMID9554879 | ChEMBL |
| EC50 | 2.2 nM | PMID15481977 | BindingDB,ChEMBL |
| EC50 | 2.45 nM | PMID9554879 | ChEMBL |
| EC50 | 2.5 nM | PMID9554879, PMID20175517, PMID15481977 | BindingDB,ChEMBL |
| EC50 | 2.6 nM | PMID15481977 | BindingDB,ChEMBL |
| EC50 | 3.0 nM | PMID15481977 | BindingDB |
| EC50 | 3.0 nM | PMID15481977 | ChEMBL |
| EC50 | 3.2 nM | PMID15481977 | BindingDB,ChEMBL |
| EC50 | 3.6 nM | PMID15481977 | BindingDB,ChEMBL |
| EC50 | 3.9 nM | PMID15481977 | BindingDB,ChEMBL |
| EC50 | 4.7 nM | PMID15481977 | BindingDB,ChEMBL |
| EC50 | 4.9 nM | PMID15481977 | BindingDB,ChEMBL |
| EC50 | 5.9 nM | PMID15481977 | BindingDB,ChEMBL |
| EC50 | 6.0 nM | PMID15481977 | BindingDB |
| EC50 | 6.0 nM | PMID15481977 | ChEMBL |
| EC50 | 6.4 nM | PMID11985476 | BindingDB,ChEMBL |
| EC50 | 7.8 nM | PMID15481977 | BindingDB,ChEMBL |
| EC50 | 8.6 nM | PMID15481977 | BindingDB,ChEMBL |
| EC50 | 9.2 nM | PMID15481977 | BindingDB,ChEMBL |
| EC50 | 9.8 nM | PMID15481977 | BindingDB,ChEMBL |
| EC50 | 10.0 nM | PMID9554879 | BindingDB |
| EC50 | 10.3 nM | PMID20192270 | BindingDB,ChEMBL |
| EC50 | 10.4 nM | PMID9554879 | ChEMBL |
| EC50 | 10.9 nM | PMID9554879 | ChEMBL |
| EC50 | 11.0 nM | PMID9554879, PMID16539385, PMID15481977 | BindingDB,ChEMBL |
| EC50 | 11.2 nM | PMID15481977 | ChEMBL |
| EC50 | 11.7 nM | PMID15481977 | ChEMBL |
| EC50 | 12.0 nM | PMID9554879, PMID15481977 | BindingDB |
| EC50 | 12.1 nM | PMID9554879 | ChEMBL |
| EC50 | 12.3 nM | PMID17941622, PMID9554879 | ChEMBL |
| EC50 | 12.4 nM | PMID9554879 | ChEMBL |
| EC50 | 12.6 nM | PMID15481977 | ChEMBL |
| EC50 | 12.9 nM | PMID9554879 | ChEMBL |
| EC50 | 13.0 nM | PMID26447940, PMID9554879, PMID23751098, PMID15481977 | BindingDB,ChEMBL |
| EC50 | 15.0 nM | PMID15481977 | BindingDB,ChEMBL |
| EC50 | 15.9 nM | PMID15481977 | ChEMBL |
| EC50 | 16.0 nM | PMID15481977 | BindingDB,ChEMBL |
| EC50 | 17.5 nM | PMID15481977 | ChEMBL |
| EC50 | 17.7 nM | PMID9554879 | ChEMBL |
| EC50 | 18.0 nM | PMID9554879, PMID15481977 | BindingDB |
| EC50 | 18.2 nM | PMID15481977 | ChEMBL |
| EC50 | 19.0 nM | PMID9554879, PMID15481977 | BindingDB,ChEMBL |
| EC50 | 19.2 nM | PMID9554879 | ChEMBL |
| EC50 | 19.5 nM | PMID9554879 | ChEMBL |
| EC50 | 20.0 nM | PMID9554879 | BindingDB |
| EC50 | 20.7 nM | PMID15481977 | ChEMBL |
| EC50 | 21.0 nM | PMID15481977 | BindingDB |
| EC50 | 25.0 nM | PMID9554879 | BindingDB,ChEMBL |
| EC50 | 27.0 nM | PMID9554879 | BindingDB,ChEMBL |
| EC50 | 30.9 nM | PMID9554879 | ChEMBL |
| EC50 | 31.0 nM | PMID9554879 | BindingDB |
| EC50 | 36.0 nM | PMID9554879 | BindingDB,ChEMBL |
| EC50 | 53.0 nM | PMID9554879 | BindingDB,ChEMBL |
| EC50 | 66.5 nM | PMID15481977 | ChEMBL |
| EC50 | 67.0 nM | PMID15481977 | BindingDB |
| EC50 | 74.9 nM | PMID15481977 | ChEMBL |
| EC50 | 75.0 nM | PMID15481977 | BindingDB |
| EC50 | 100.0 nM | PMID15481977 | BindingDB,ChEMBL |
| EC50 | 103.0 nM | PMID9554879 | BindingDB,ChEMBL |
| EC50 | 115.0 nM | PMID9554879 | BindingDB,ChEMBL |
| EC50 | 145.0 nM | PMID15481977 | BindingDB,ChEMBL |
| EC50 | 323.0 nM | PMID9554879 | BindingDB,ChEMBL |
| EC50 | 324.0 nM | PMID9554879 | BindingDB,ChEMBL |
| EC50 | 378.0 nM | PMID9554879 | BindingDB,ChEMBL |
| EC50 | 418.0 nM | PMID15481977 | BindingDB,ChEMBL |
| EC50 | 455.0 nM | PMID15481977 | BindingDB,ChEMBL |
| EC50 | 595.0 nM | PMID15481977 | BindingDB,ChEMBL |
| EC50 | 738.0 nM | PMID9554879 | BindingDB,ChEMBL |
| EC50 | 1520.0 nM | PMID9554879 | BindingDB,ChEMBL |
| EC50 | 1580.0 nM | PMID9554879 | BindingDB,ChEMBL |
| EC50 | 1790.0 nM | PMID15481977 | BindingDB,ChEMBL |
| EC50 | 2270.0 nM | PMID9554879 | BindingDB,ChEMBL |
| EC50 | 5160.0 nM | PMID15481977 | BindingDB,ChEMBL |
| EC50 | 5290.0 nM | PMID15481977 | BindingDB,ChEMBL |
| EC50 | 14800.0 nM | PMID15481977 | BindingDB,ChEMBL |
| EC50 | 17200.0 nM | PMID15481977 | BindingDB,ChEMBL |
| IC50 | <10000.0 nM | PMID15481977 | BindingDB,ChEMBL |
| IC50 | 22.0 nM | PMID15481977 | BindingDB,ChEMBL |
| IC50 | 24.0 nM | PMID15481977 | BindingDB,ChEMBL |
| IC50 | 40.0 nM | PMID15481977 | BindingDB,ChEMBL |
| IC50 | 100.0 - 3981.07 nM | PMID9154346, PMID12391289 | IUPHAR |
| IC50 | 150.0 nM | PMID15481977 | BindingDB,ChEMBL |
| IC50 | 260.0 nM | PMID15481977 | BindingDB,ChEMBL |
| IC50 | 360.0 nM | PMID15481977 | BindingDB,ChEMBL |
| Kd | 49.0 nM | PMID11502873 | IUPHAR |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417