

You can:
| Name | Glucagon receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | GCGR |
| Synonym | GL-R GGR GR glucagon receptor |
| Disease | Diabetes Type 2 diabetes Non-insulin dependent diabetes Obesity; Diabetes Type 1 diabetes [ Show all ] |
| Length | 477 |
| Amino acid sequence | MPPCQPQRPLLLLLLLLACQPQVPSAQVMDFLFEKWKLYGDQCHHNLSLLPPPTELVCNRTFDKYSCWPDTPANTTANISCPWYLPWHHKVQHRFVFKRCGPDGQWVRGPRGQPWRDASQCQMDGEEIEVQKEVAKMYSSFQVMYTVGYSLSLGALLLALAILGGLSKLHCTRNAIHANLFASFVLKASSVLVIDGLLRTRYSQKIGDDLSVSTWLSDGAVAGCRVAAVFMQYGIVANYCWLLVEGLYLHNLLGLATLPERSFFSLYLGIGWGAPMLFVVPWAVVKCLFENVQCWTSNDNMGFWWILRFPVFLAILINFFIFVRIVQLLVAKLRARQMHHTDYKFRLAKSTLTLIPLLGVHEVVFAFVTDEHAQGTLRSAKLFFDLFLSSFQGLLVAVLYCFLNKEVQSELRRRWHRWRLGKVLWEERNTSNHRASSSPGHGPPSKELQFGRGGGSQDSSAETPLAGGLPRLAESPF |
| UniProt | P47871 |
| Protein Data Bank | 5yqz, 5xez, 5ee7, 5xf1 |
| GPCR-HGmod model | P47871 |
| 3D structure model | This structure is from PDB ID 5yqz. |
| BioLiP | BL0379634,BL0379635, BL0402227, BL0379636,BL0379637, BL0343437 |
| Therapeutic Target Database | T60182, T87875 |
| ChEMBL | CHEMBL1985 |
| IUPHAR | 251 |
| DrugBank | N/A |
| Name | CHEMBL386446 |
|---|---|
| Molecular formula | C30H29Cl2N3O4 |
| IUPAC name | 3-[[4-[[4-(cyclohexen-1-yl)-N-[(3,5-dichlorophenyl)carbamoyl]anilino]methyl]benzoyl]amino]propanoic acid |
| Molecular weight | 566.479 |
| Hydrogen bond acceptor | 4 |
| Hydrogen bond donor | 3 |
| XlogP | 5.9 |
| Synonyms | SCHEMBL2652197 3-(4-((1-(4-cyclohexenylphenyl)-3-(3,5-dichlorophenyl)ureido)methyl)benzamido)propanoic acid BDBM50202031 LS-193209 3-{4-[1-(4-Cyclohex-1-enylphenyl)-3-(3,5-dichlorophenyl)ureidomethyl]benzoylamino}propionicacid |
| Inchi Key | VHURVYZHODPCCB-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C30H29Cl2N3O4/c31-24-16-25(32)18-26(17-24)34-30(39)35(27-12-10-22(11-13-27)21-4-2-1-3-5-21)19-20-6-8-23(9-7-20)29(38)33-15-14-28(36)37/h4,6-13,16-18H,1-3,5,14-15,19H2,(H,33,38)(H,34,39)(H,36,37) |
| PubChem CID | 10257357 |
| ChEMBL | CHEMBL386446 |
| IUPHAR | N/A |
| BindingDB | 50202031 |
| DrugBank | N/A |
Structure | 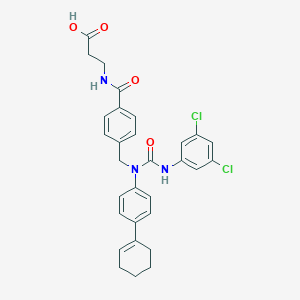 |
| Lipinski's druglikeness | This ligand is heavier than 500 daltons. This ligand has a partition coefficient log P greater than 5. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| IC50 | 12.0 nM | PMID18707090 | BindingDB,ChEMBL |
| IC50 | 27.0 nM | PMID17201415 | BindingDB,ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417